|
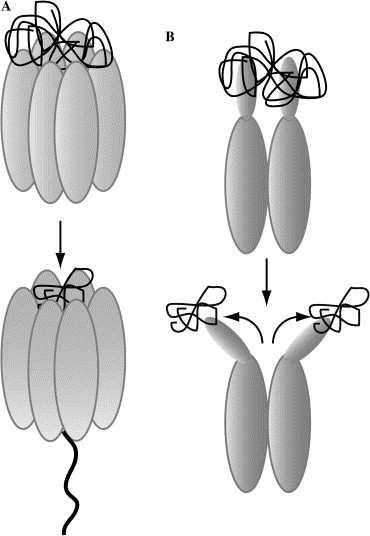
Clp Protease
Endopeptidase Clp (, ''endopeptidase Ti'', ''caseinolytic protease'', ''protease Ti'', ''ATP-dependent Clp protease'', '' ClpP'', ''Clp protease''). This enzyme catalyses the following chemical reaction : Hydrolysis of proteins to small peptides in the presence of ATP and Mg2+. This bacterial enzyme contains subunits of two types, ClpP, with peptidase activity, and the protein ClpA, with AAA+ ATPase activity. ClpP and ClpA are not evolutionarily related. A fully assembled Clp protease complex has a barrel-shaped structure in which two stacked heptameric ring of proteolytic subunits ( ClpP or ClpQ) are either sandwiched between two rings or single-caped by one ring of hexameric ATPase-active chaperon subunits (ClpA, ClpC, ClpE, ClpX, ClpY, or others). ClpXP is presented in almost all bacteria while ClpA is found in the Gram-negative bacteria, ClpC in Gram-Positive bacteria and cyanobacteria. ClpAP, ClpXP and ClpYQ coexist in ''E. coli'' while only ClpXP complex in present in h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heat Shock Protein
Heat shock proteins (HSPs) are a family of proteins produced by cells in response to exposure to stressful conditions. They were first described in relation to heat shock, but are now known to also be expressed during other stresses including exposure to cold, UV light and during wound healing or tissue remodeling. Many members of this group perform chaperone functions by stabilizing new proteins to ensure correct folding or by helping to refold proteins that were damaged by the cell stress. This increase in expression is transcriptionally regulated. The dramatic upregulation of the heat shock proteins is a key part of the heat shock response and is induced primarily by heat shock factor (HSF). HSPs are found in virtually all living organisms, from bacteria to humans. Heat shock proteins are named according to their molecular weight. For example, Hsp60, Hsp70 and Hsp90 (the most widely studied HSPs) refer to families of heat shock proteins on the order of 60, 70 and 90 kilo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endopeptidase
Endopeptidase or endoproteinase are proteolytic peptidases that break peptide bonds of nonterminal amino acids (i.e. within the molecule), in contrast to exopeptidases, which break peptide bonds from end-pieces of terminal amino acids. For this reason, endopeptidases cannot break down peptides into monomers, while exopeptidases can break down proteins into monomers. A particular case of endopeptidase is the oligopeptidase, whose substrates are oligopeptides instead of proteins. They are usually very specific for certain amino acids. Examples of endopeptidases include: * Trypsin - cuts after Arg or Lys, unless followed by Pro. Very strict. Works best at pH 8. * Chymotrypsin - cuts after Phe, Trp, or Tyr, unless followed by Pro. Cuts more slowly after His, Met or Leu. Works best at pH 8. * Elastase - cuts after Ala, Gly, Ser, or Val, unless followed by Pro. * Thermolysin - cuts ''before'' Ile, Met, Phe, Trp, Tyr, or Val, unless ''preceded'' by Pro. Sometimes cuts after Ala, Asp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
T6SS
The type VI secretion system (T6SS) is one of the bacterial secretion systems, membrane protein complexes, used by a wide range of gram-negative bacteria to transport effectors. Effectors are moved from the interior of a bacterial cell, across the membrane into an adjacent target cell. While often reported that the T6SS was discovered in 2006 by researchers studying the causative agent of cholera, ''Vibrio cholerae'', the first study demonstrating that T6SS genes encode a protein export apparatus was actually published in 2004, in a study of protein secretion by the fish pathogen '' Edwardsiella tarda''. Since then, it is estimated that at least a quarter of all pathogenic and non-pathogenic proteobacterial genomes encode for a T6SS, including pathogens of animals, plants, and humans, as well as soil, environmental or marine bacteria. Genes encoding for the T6SSs are commonly found chromosomally, but can also be harboured in mobile genetic elements and on plasmids mediating thei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nanometer
330px, Different lengths as in respect to the Molecule">molecular scale. The nanometre (international spelling as used by the International Bureau of Weights and Measures; SI symbol: nm), or nanometer (American spelling Despite the various list of dialects of English, English dialects spoken from country to country and within different regions of the same country, there are only slight regional variations in English orthography, the two most notable variati ...), is a units of measurement, unit of length in the International System of Units (SI), equal to one billionth (short scale) or one thousand million (long scale) of a metre, meter (0.000000001 m) and to 1000 picometres. One nanometre can be expressed in scientific notation as 1 × 10−9 m and as m. History The nanometre was formerly known as the "''millimicrometre''" – or, more commonly, the "''millimicron''" for short – since it is of a micrometre, micrometer. It was often de ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hsp70
The 70 kilodalton heat shock proteins (Hsp70s or DnaK) are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms and play crucial roles in the development of cancer, neurodegeneration, apoptosis, regulating sleep, and much more. Intracellularly localized Hsp70s are an important part of the cell's machinery for protein folding, performing chaperoning functions, and helping to protect cells from the adverse effects of physiological stresses. Additionally, membrane-bound Hsp70s have been identified as a potential target for cancer therapies and their extracellularly localized counterparts have been identified as having both membrane-bound and membrane-free structures. There is lot of potential in the Hsp70 protein as a key therapeutic target for developing new drugs for the treatment of sleep disorders, cancer, neurodegeneration, and other related pathological conditions. Discovery Members of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hsp104
Hsp104 is a heat-shock protein. It is known to reverse toxicity of mutant α-synuclein, TDP-43, FUS (gene), FUS, and TAF15 in yeast cells. Conserved in prokaryotes (ClpB), fungi, plants and as well as animal mitochondria, there is yet to see hsp104 in multicellular animals. Hsp104 is classified as a. AAA+ ATPases and a subgroup of Hsp100/Clp, because of the usage of Atp hydrolysis for structural modulation of other proteins. Hsp104 is not needed for normal cell growth but when exposed to stress there is an increasing amount. Removing the aggregates without the hsp104 is insufficient there highlighting the importance of this heat shock protein and its interactions. Structure Hsp104 monomer is composed of two NBDs (Nucleotide Binding Sites) NBD1 and NBD2 which communicate through allosteric communication. Located on the C-terminus of NBD1 there are around 125 residues that link both NBDs. In hsp104 NBD1 is where ATP hydrolysis occurs, NBD2 C-terminus is shown to express the configu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CLPB
Caseinolytic peptidase B protein homolog (''CLPB''), also known as Skd3, is a mitochondrial AAA ATPase chaperone that in humans is encoded by the gene ''CLPB'', which encodes an adenosine triphosphate-(ATP) dependent chaperone. Skd3 is localized in mitochondria and widely expressed in human tissues. High expression in adult brain and low expression in granulocyte is found. It is a potent protein disaggregase that chaperones the mitochondrial intermembrane space. Mutations in the ''CLPB'' gene could cause autosomal recessive metabolic disorder with intellectual disability/developmental delay, congenital neutropenia, progressive brain atrophy, movement disorder, cataracts, and 3-methylglutaconic aciduria 3-Methylglutaconic aciduria (MGA) is any of at least five metabolic disorders that impair the body's ability to make energy in the mitochondria. As a result of this impairment, 3-methylglutaconic acid and 3-methylglutaric acid build up and can be .... Recently, heterozygous, domin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteasome
Proteasomes are essential protein complexes responsible for the degradation of proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases. Proteasomes are found inside all eukaryotes and archaea, and in some bacteria. In eukaryotes, proteasomes are located both in the nucleus and in the cytoplasm. The proteasomal degradation pathway is essential for many cellular processes, including the cell cycle, the regulation of gene expression, and responses to oxidative stress. The importance of proteolytic degradation inside cells and the role of ubiquitin in proteolytic pathways was acknowledged in the award of the 2004 Nobel Prize in Chemistry to Aaron Ciechanover, Avram Hershko and Irwin Rose. The core 20S proteasome (blue in the adjacent figure) is a cylindrical, compartmental protein complex of four stacked rings forming a central pore. Each ring is composed of seven individual proteins. The inner two rings a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HslVU
The heat shock proteins HslV and HslU (HslVU complex; also known as ClpQ and ClpY respectively, or ClpQY) are expressed in many bacteria such as ''E. coli'' in response to cell stress.Ramachandran R, Hartmann C, Song HK, Huber R, Bochtler M. (2002). Functional interactions of HslV (ClpQ) with the ATPase HslU (ClpY). ''Proc Natl Acad Sci USA'' 99(11):7396-401. The hslV protein is a protease and the hslU protein is an ATPase; the two form a symmetric assembly of four stacked rings, consisting of an hslV dodecamer bound to an hslU hexamer, with a central pore in which the protease and ATPase active sites reside. The hslV protein degrades unneeded or damaged proteins only when in complex with the hslU protein in the ATP-bound state. HslV is thought to resemble the hypothetical ancestor of the proteasome, a large protein complex specialized for regulated degradation of unneeded proteins in eukaryotes, many archaea, and a few bacteria. HslV bears high similarity to core subunits of pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalysis
Catalysis () is the increase in rate of a chemical reaction due to an added substance known as a catalyst (). Catalysts are not consumed by the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quickly, very small amounts of catalyst often suffice; mixing, surface area, and temperature are important factors in reaction rate. Catalysts generally react with one or more reactants to form intermediates that subsequently give the final reaction product, in the process of regenerating the catalyst. The rate increase occurs because the catalyst allows the reaction to occur by an alternative mechanism which may be much faster than the noncatalyzed mechanism. However the noncatalyzed mechanism does remain possible, so that the total rate (catalyzed plus noncatalyzed) can only increase in the presence of the catalyst and never decrease. Catalysis may be classified as either homogeneous, whose components are dispersed in the same phase (usual ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |