|
Circulating Tumor DNA
Circulating tumor DNA (ctDNA) is tumor-derived fragmented DNA in the bloodstream that is not associated with cells. ctDNA should not be confused with cell-free DNA ( cfDNA), a broader term which describes DNA that is freely circulating in the bloodstream, but is not necessarily of tumor origin. Because ctDNA may reflect the entire tumor genome, it has gained traction for its potential clinical utility; " liquid biopsies" in the form of blood draws may be taken at various time points to monitor tumor progression throughout the treatment regimen. Recent studies have laid the foundation for inferring gene expression from cfDNA (and ctDNA), with EPIC-seq emerging as a notable advancement. This method has substantially raised the bar for the noninvasive inference of expression levels of individual genes, thereby augmenting the assay's applicability in disease characterization, histological classification, and monitoring treatment efficacy. ctDNA originates directly from the tumor or f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bert Vogelstein
Bert Vogelstein (born 1949) is director of the Ludwig Center, Clayton Professor of Oncology and Pathology and a Howard Hughes Medical Institute investigator at The Johns Hopkins Medical School and Sidney Kimmel Comprehensive Cancer Center. A pioneer in the field of cancer genomics, his studies on colorectal cancers revealed that they result from the sequential accumulation of mutations in oncogenes and tumor suppressor genes. These studies now form the paradigm for modern cancer research and provided the basis for the notion of the somatic evolution of cancer. Research In the 1980s, Vogelstein developed new experimental approaches to study human tumors. His studies of various stages of colorectal cancers led him to propose a specific model for human tumorigenesis in 1988. In particular, he suggested that "cancer is caused by sequential mutations of specific oncogenes and tumor suppressor genes". The first tumor suppressor gene validating this hypothesis was that encoding p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BEAMing
In physics, relativistic beaming (also known as Doppler beaming, Doppler boosting, or the headlight effect) is the process by which relativistic effects modify the apparent luminosity of emitting matter that is moving at speeds close to the speed of light. In an astronomical context, relativistic beaming commonly occurs in two oppositely-directed relativistic jets of plasma that originate from a central compact object that is accreting matter. Accreting compact objects and relativistic jets are invoked to explain x-ray binaries, gamma-ray bursts, and, on a much larger scale, (AGN) active galactic nuclei (of which quasars are a particular variety). Beaming affects the apparent brightness of a moving object. Consider a cloud of gas moving relative to the observer and emitting electromagnetic radiation. If the gas is moving towards the observer, it will be brighter than if it were at rest, but if the gas is moving away, it will appear fainter. The magnitude of the effect i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Digital Polymerase Chain Reaction
Digital polymerase chain reaction (digital PCR, DigitalPCR, dPCR, or dePCR) is a biotechnological refinement of conventional polymerase chain reaction methods that can be used to directly quantify and clonally amplify nucleic acids strands including DNA, cDNA, or RNA. The key difference between dPCR and qPCR lies in the method of measuring nucleic acids amounts, with the former being a more precise method than PCR, though also more prone to error in the hands of inexperienced users. PCR carries out one reaction per single sample. dPCR also carries out a single reaction within a sample, however the sample is separated into a large number of partitions and the reaction is carried out in each partition individually. This separation allows a more reliable collection and sensitive measurement of nucleic acid amounts. The method has been demonstrated as useful for studying variations in gene sequences—such as copy number variants and point mutations. Principles The polymerase cha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research and forensic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, a Guanine, G nucleotide present at a specific location in a reference genome may be replaced by an Adenine, A in a minority of individuals. The two possible nucleotide variations of this SNP – G or A – are called alleles. SNPs can help explain differences in susceptibility to a wide range of diseases across a population. For example, a common SNP in the Factor H, CFH gene is associated with increased risk of age-related macular degeneration. Differences in the severity of an illness or response to treatments may also be manifestations of genetic variations caused by SNPs. For example, two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bisulfite Sequencing
Bisulfite sequencing (also known as bisulphite sequencing) is the use of bisulfite treatment of DNA before routine sequencing to determine the pattern of methylation. DNA methylation was the first discovered epigenetic mark, and remains the most studied. In animals it predominantly involves the addition of a methyl group to the carbon-5 position of cytosine residues of the dinucleotide CpG, and is implicated in repression of transcriptional activity. Treatment of DNA with bisulfite converts cytosine residues to uracil, but leaves 5-methylcytosine residues unaffected. Therefore, DNA that has been treated with bisulfite retains only methylated cytosines. Thus, bisulfite treatment introduces specific changes in the DNA sequence that depend on the methylation status of individual cytosine residues, yielding single-nucleotide resolution information about the methylation status of a segment of DNA. Various analyses can be performed on the altered sequence to retrieve this informatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CpG Islands
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG islands. Cytosines in CpG dinucleotides can be methylated to form 5-methylcytosines. Enzymes that add a methyl group are called DNA methyltransferases. In mammals, 70% to 80% of CpG cytosines are methylated. Methylating the cytosine within a gene can change its expression, a mechanism that is part of a larger field of science studying gene regulation that is called epigenetics. Methylated cytosines often mutate to thymines. In humans, about 70% of promoters located near the transcription start site of a gene (proximal promoters) contain a CpG island. CpG characteristics Definition ''CpG'' is shorthand for ''5'—C—phosphate—G—3' '', that is, cytosine and guanine separated by only one phosphate group; phosphate links any two n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylation typically acts to repress gene Transcription (genetics), transcription. In mammals, DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis. As of 2016, two nucleobases have been found on which natural, enzymatic DNA methylation takes place: adenine and cytosine. The modified bases are N6-methyladenine,D. B. Dunn, J. D. Smith: "The occurrence of 6-methylaminopurine in deoxyribonucleic acids". In: ''Biochem J.'' 68(4), Apr 1958, S. 627–636. [//www.ncbi.nlm.nih.gov/pubmed/13522672?dopt=Abstract PMID 13522672]. . 5-methylcytosineB. F. Vanyushin, S. G. Tkacheva, A. N. Belozers ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetics
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional (DNA sequence based) genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological traits may result from environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Further, non-coding RNA sequences have been shown to play a key role in the r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
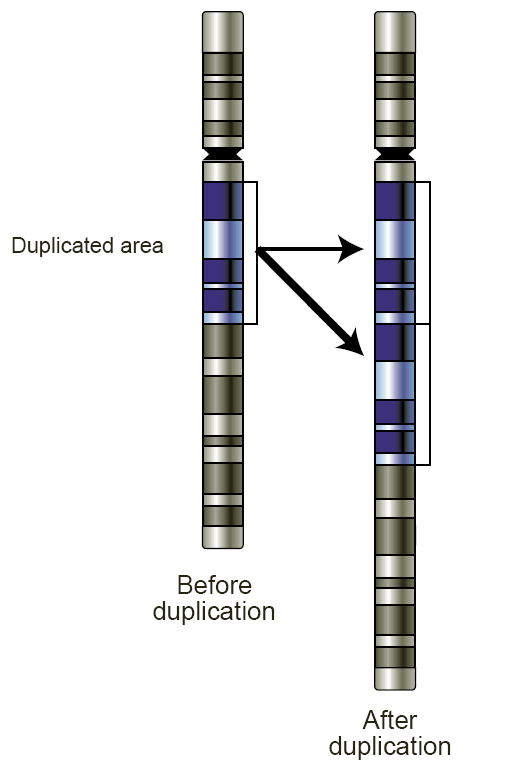
Copy-number Variation
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammals, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype. Copy number variations can be generally categorized into two main groups: short repeats and long repeats. However, there are no clear boundaries between the two groups and the classification depends on the nature of the loci of interest. Short repeats include mainly dinucleotide repeats (two repeating nucleotides e.g. A-C-A-C-A-C...) and trinucleotide repeats. Lon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |