|
Aptamer
Aptamers are oligomers of artificial ssDNA, RNA, Xeno nucleic acid, XNA, or peptide that ligand, bind a specific target molecule, or family of target molecules. They exhibit a range of affinities (Dissociation constant, KD in the pM to μM range), with variable levels of Antitarget, off-target binding and are sometimes classified as antibody mimetic, chemical antibodies. Aptamers and antibodies can be used in many of the same applications, but the nucleic acid-based structure of aptamers, which are mostly oligonucleotides, is very different from the amino acid-based structure of antibodies, which are proteins. This difference can make aptamers a better choice than antibodies for some purposes (see #Antibody_replacement, antibody replacement). Aptamers are used in biological lab research and medical tests. If multiple aptamers are combined into a single assay, they can proteomics, measure large numbers of different proteins in a sample. They can be used to biomarker discovery, id ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Therapeutics
RNA therapeutics are a new class of medications based on RNA, ribonucleic acid (RNA). Research has been working on clinical use since the 1990s, with significant success in cancer therapy in the early 2010s. In 2020 and 2021, mRNA vaccines have been developed globally for use in combating the coronavirus disease 2019, coronavirus disease (COVID-19 pandemic). The Pfizer–BioNTech COVID-19 vaccine was the first mRNA vaccine approved by a Regulation of therapeutic goods, medicines regulator, followed by the Moderna COVID-19 vaccine, and others. The main types of RNA therapeutics are those based on messenger RNA (mRNA), antisense RNA (asRNA), RNA interference (RNAi), RNA activation (RNAa) and RNA aptamers. Of the four types, mRNA-based therapy is the only type which is based on triggering Protein biosynthesis, synthesis of proteins within cells, making it particularly useful in vaccine development. Antisense RNA is complementary to coding mRNA and is used to trigger mRNA inactivation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteomics
Proteomics is the large-scale study of proteins. Proteins are vital macromolecules of all living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body. The proteome is the entire set of proteins produced or modified by an organism or system. Proteomics enables the identification of ever-increasing numbers of proteins. This varies with time and distinct requirements, or stresses, that a cell or organism undergoes. Proteomics is an interdisciplinary domain that has benefited greatly from the genetic information of various genome projects, including the Human Genome Project. It covers the exploration of proteomes from the overall level of protein composition, structure, and activity, and is an important component of function ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Modified-release Dosage
Modified-release dosage is a mechanism that (in contrast to immediate-release dosage) delivers a drug with a delay after its administration (delayed-release dosage) or for a prolonged period of time (extended-release R, XR, XLdosage) or to a specific target in the body (targeted-release dosage).Pharmaceutics: Drug Delivery and Targeting p. 7-13 Sustained-release dosage forms are dosage forms designed to release (liberate) a drug at a predetermined rate in order to maintain a constant drug concentration for a specific period of time ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antibody Replacement
An antibody (Ab) or immunoglobulin (Ig) is a large, Y-shaped protein belonging to the immunoglobulin superfamily which is used by the immune system to identify and neutralize antigens such as bacteria and viruses, including those that cause disease. Each individual antibody recognizes one or more specific antigens, and antigens of virtually any size and chemical composition can be recognized. Antigen literally means "antibody generator", as it is the presence of an antigen that drives the formation of an antigen-specific antibody. Each of the branching chains comprising the "Y" of an antibody contains a paratope that specifically binds to one particular epitope on an antigen, allowing the two molecules to bind together with precision. Using this mechanism, antibodies can effectively "tag" the antigen (or a microbe or an infected cell bearing such an antigen) for attack by cells of the immune system, or can neutralize it directly (for example, by blocking a part of a virus that i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
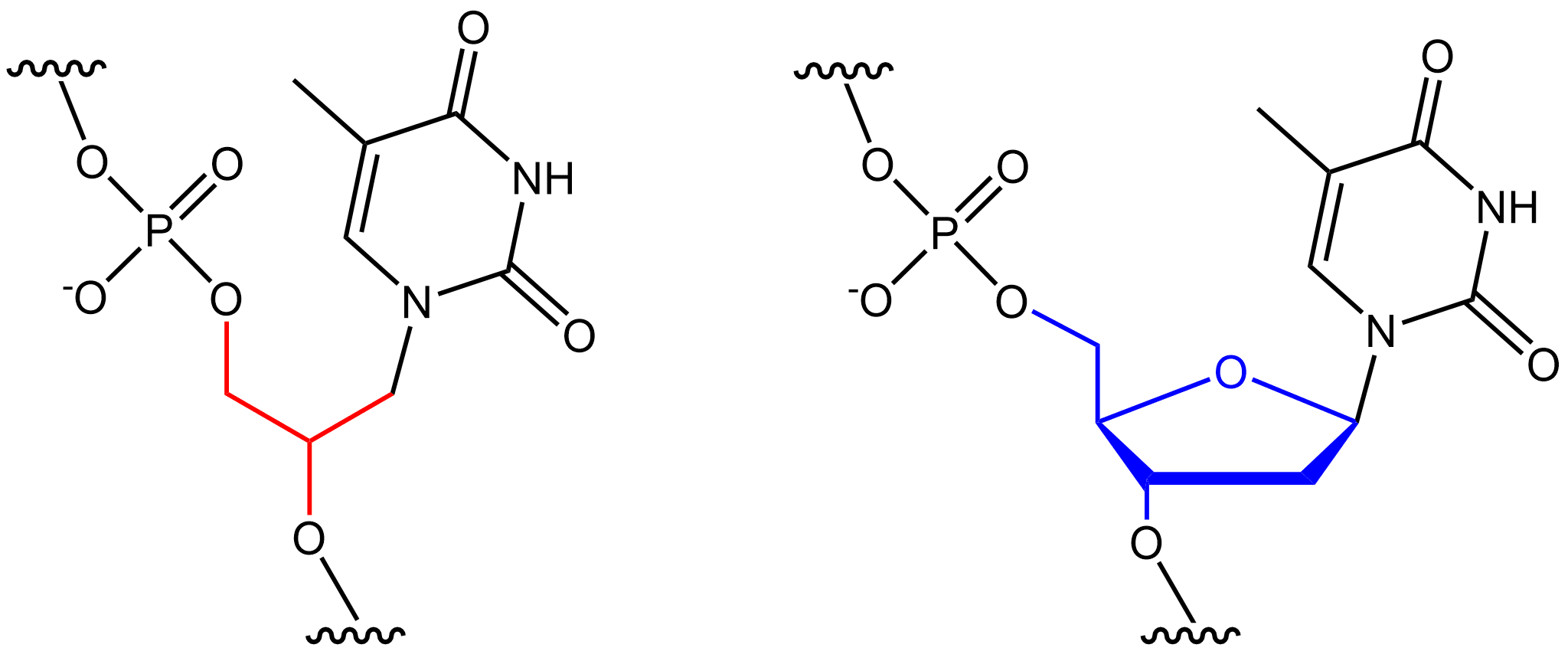
Oligonucleotides
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small fragments of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression (e.g. microRNA), or are degradation intermediates derived from the breakdown of larger nucleic acid molecules. Oligonucleotides are characterized by the sequence of nucleotide residues that make up the entire molecule. The length of the oligonucleotide is usually denoted by " -mer" (from Greek ''meros'', "part"). For example, an oligonucleotide of six nucleotides (nt) is a hexamer, while one of 25 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antibody Mimetic
Antibody mimetics are organic compounds that, like antibodies, can specifically bind antigens, but that are not structurally related to antibodies. They are usually artificial peptides or proteins with a molar mass of about 3 to 20 kDa. (Antibodies are ~150 kDa.) Nucleic acids and small molecules are sometimes considered antibody mimetics as well, but not artificial antibodies, Fragment antigen-binding, antibody fragments and fusion proteins composed from these. Common advantages over antibodies are better solubility, tissue penetration, stability towards heat and enzymes, and comparatively low production costs. Antibody mimetics are being developed as therapeutic and Medical diagnosis, diagnostic agents. Examples See also *Protein mimetic *Optimer Ligand References {{Engineered antibodies Antibody mimetics, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Xeno Nucleic Acid
Xenonucleic acids (XNAs) are synthetic nucleic acid analogues that are engineered with structurally distinct components, such as alternative nucleosides, sugars, or backbones. XNAs have fundamentally different properties from endogenous nucleic acids, enabling different specialized applications, such as therapeutics, probes, or functional molecules. For instance, peptide nucleic acids, the backbones of which are made up of repeating aminoethylglycine units, are extremely stable and resistant to degradation by nucleases because they are not recognised. The same nucleobases can be used to store genetic information and interact with DNA, RNA, or other XNA bases, but the different backbone gives the compound different properties. Their altered chemical structure means they cannot be processed by naturally occurring cellular processes. For instance, natural DNA polymerases cannot read and duplicate the alien information, thus the genetic information stored in XNA is invisible to DNA- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quadrillion
Depending on context (e.g. language, culture, region), some large numbers have names that allow for describing large quantities in a textual form; not mathematical. For very large values, the text is generally shorter than a decimal numeric representation although longer than scientific notation. Two naming scales for large numbers have been used in English and other European languages since the early modern era: the long and short scales. Most English variants use the short scale today, but the long scale remains dominant in many non-English-speaking areas, including continental Europe and Spanish-speaking countries in Latin America. These naming procedures are based on taking the number ''n'' occurring in 103''n''+3 (short scale) or 106''n'' (long scale) and concatenating Latin roots for its units, tens, and hundreds place, together with the suffix ''-illion''. Names of numbers above a trillion are rarely used in practice; such large numbers have practical usage primarily in th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
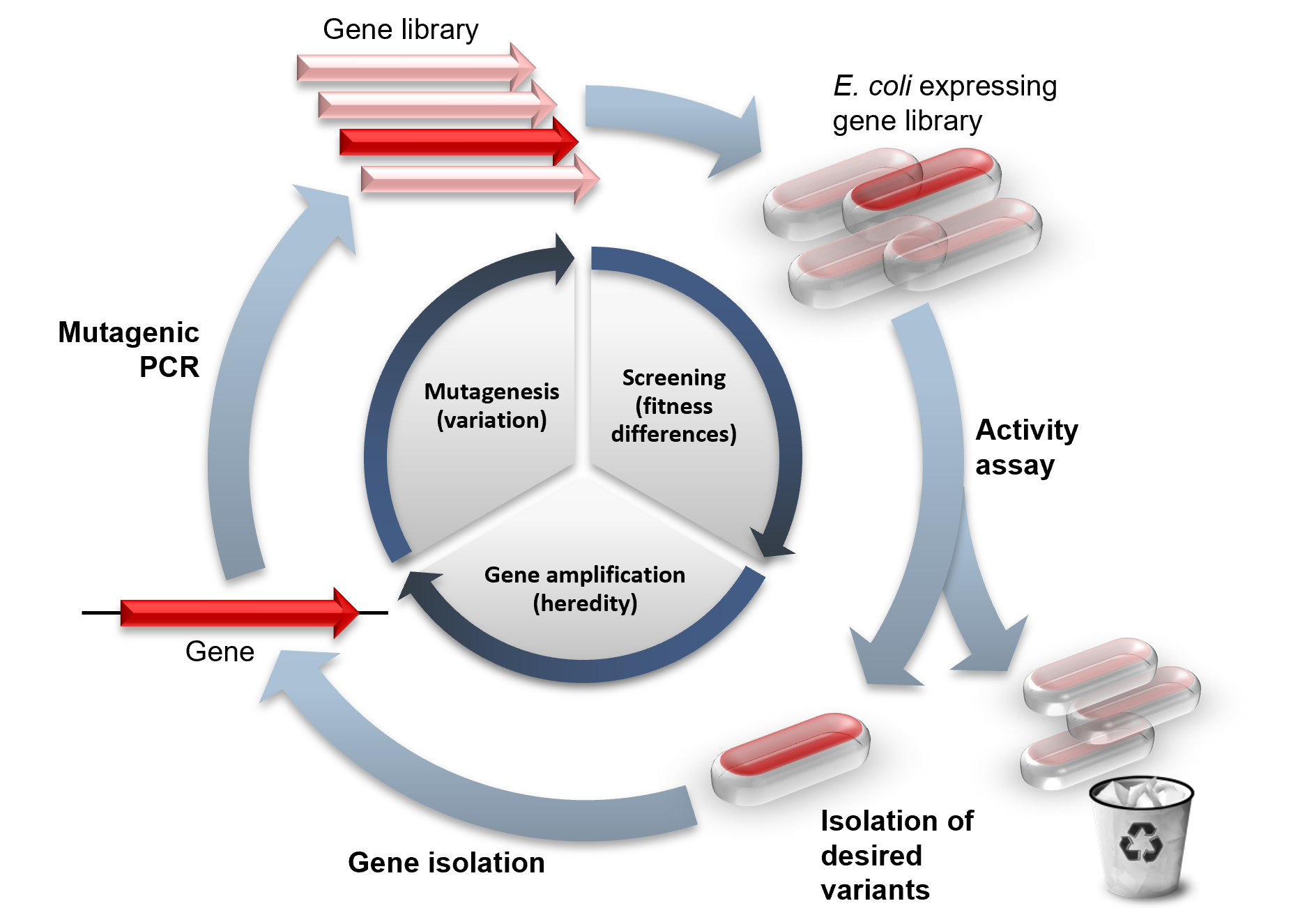
Directed Evolution
Directed evolution (DE) is a method used in protein engineering that mimics the process of natural selection to steer proteins or nucleic acids toward a user-defined goal. It consists of subjecting a gene to iterative rounds of mutagenesis (creating a library of variants), selection (expressing those variants and isolating members with the desired function) and amplification (generating a template for the next round). It can be performed ''in vivo'' (in living organisms), or ''in vitro'' (in cells or free in solution). Directed evolution is used both for protein engineering as an alternative to rationally designing modified proteins, as well as for experimental evolution studies of fundamental evolutionary principles in a controlled, laboratory environment. History Directed evolution has its origins in the 1960s with the evolution of RNA molecules in the "Spiegelman's Monster" experiment. The concept was extended to protein evolution via evolution of bacteria under selec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Natural Selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the Heredity, heritable traits characteristic of a population over generations. Charles Darwin popularised the term "natural selection", contrasting it with selective breeding, artificial selection, which is intentional, whereas natural selection is not. Genetic diversity, Variation of traits, both Genotype, genotypic and phenotypic, exists within all populations of organisms. However, some traits are more likely to facilitate survival and reproductive success. Thus, these traits are passed the next generation. These traits can also become more Allele frequency, common within a population if the environment that favours these traits remains fixed. If new traits become more favoured due to changes in a specific Ecological niche, niche, microevolution occurs. If new traits become more favoured due to changes in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligonucleotide Synthesis
Oligonucleotide synthesis is the chemical synthesis of relatively short fragments of nucleic acids with defined chemical structure (sequence). The technique is extremely useful in current laboratory practice because it provides a rapid and inexpensive access to custom-made oligonucleotides of the desired sequence. Whereas enzymes synthesize DNA and RNA only in a 5' to 3' direction, chemical oligonucleotide synthesis does not have this limitation, although it is most often carried out in the opposite, 3' to 5' direction. Currently, the process is implemented as solid-phase synthesis using phosphoramidite method and phosphoramidite building blocks derived from protected 2'-deoxynucleosides ( dA, dC, dG, and T), ribonucleosides ( A, C, G, and U), or chemically modified nucleosides, e.g. LNA or BNA. To obtain the desired oligonucleotide, the building blocks are sequentially coupled to the growing oligonucleotide chain in the order required by the sequence of the product (s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
In Vitro
''In vitro'' (meaning ''in glass'', or ''in the glass'') Research, studies are performed with Cell (biology), cells or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in biology and its subdisciplines are traditionally done in labware such as test tubes, flasks, Petri dishes, and microtiter plates. Studies conducted using components of an organism that have been isolated from their usual biological surroundings permit a more detailed or more convenient analysis than can be done with whole organisms; however, results obtained from ''in vitro'' experiments may not fully or accurately predict the effects on a whole organism. In contrast to ''in vitro'' experiments, ''in vivo'' studies are those conducted in living organisms, including humans, known as clinical trials, and whole plants. Definition ''In vitro'' (Latin language, Latin for "in glass"; often not italicized in English usage) studies are conducted ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |