|
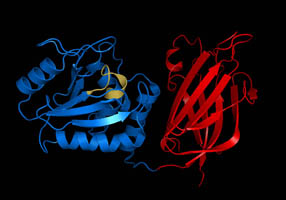
Anti-sigma Factors
Introduction Anti-sigma factors are small proteins that bind to sigma factors and inhibit transcriptional activity in regulating prokaryote gene expression. Anti-sigma factors have both a sigma-binding domain and a sensory/signaling domain; this allows them to respond to signals inside and outside the cell. Anti-sigma factors have been found in several bacteria, including ''Escherichia coli'' and ''Salmonella'', and viruses such as the T4 bacteriophage. Anti-sigma factors have an antagonistic effect on sigma factors. Each sigma factor has an associated anti-sigma factor that regulates it. These anti-sigma factors are divided into cytoplasmic-bound anti-sigma factors and inner membrane-bound anti-sigma factors. The differences in these sigma factors are where in the cell they are bound. Cytoplasmic-bound anti-sigma factors include FlgM, DnaK, RssB, and HscC. Inner membrane-bound anti-sigma factors, also called extra-cytoplasmic function (ECF) anti-sigma factors, include FecR and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metabolic reactions, DNA replication, Cell signaling, responding to stimuli, providing Cytoskeleton, structure to cells and Fibrous protein, organisms, and Intracellular transport, transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the Nucleic acid sequence, nucleotide sequence of their genes, and which usually results in protein folding into a specific Protein structure, 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called pep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteolysis
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Protein degradation is a major regulatory mechanism of gene expression and contributes substantially to shaping mammalian proteomes. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion. Proteolysis in organisms serves many purposes; for example, digestive enzymes break down proteins in food to provide amino acids for the organism, while proteolytic processing of a polypeptide chain after its synthesis may be necessary for the production of an active protein. It is also important in the regulation of some physiological and cellular processes including apoptosis, as well as preventing the accumulation of unwanted or misfolded proteins in cells. Consequently, abnormality in the regulation of proteolysis can cause diseases. Proteolysis can also ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriophage T4
Escherichia virus T4 is a species of bacteriophages that infect ''Escherichia coli'' bacteria. It is a double-stranded DNA virus in the subfamily '' Tevenvirinae'' of the family '' Straboviridae''. T4 is capable of undergoing only a lytic life cycle and not the lysogenic life cycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains (or isolates), Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6. Use in research Dating back to the 1940s and continuing today, T-even phages are considered the best studied model organisms. Model organisms are usually required to be simple with as few as five genes. Yet, T-even phages are in fact among the largest and highest complexity virus, in which these phage's genetic information is made up of around 300 genes. Coincident with their complexity, T-even viruses were found to have the unusual base hydroxymethylcytosine (HMC) in place of the nucleic acid base ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Audrey Stevens' Inhibtor
Audrey () is a feminine given name. It is rarely a masculine given name. Audrey is the Anglo-Norman form of the Anglo-Saxon name ''Æðelþryð'', composed of the elements '' æðel'' "noble" and '' þryð'' "strength". The literal definition of the word is “noble strength” or “strength from nobility”. The Anglo-Norman form of the name was applied to Saint Audrey (died 679), also known by the historical form of her name as Saint Æthelthryth. The same name also survived into the modern period in its Anglo-Saxon form, as ''Etheldred'', e.g. Etheldred Benett (1776–1845). In the 17th century, the name of ''Saint Audrey'' gave rise to the adjective ''tawdry'' "cheap and pretentious; cheaply adorned". The lace necklaces sold to pilgrims to Saint Audrey fell out of fashion in the 17th century, and so tawdry was reinterpreted as meaning cheap or vulgar. As a consequence, use of the name declined, but it was revived in the 19th century. Popularity of the name in the United St ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RpoE
The gene ''rpoE'' (RNA polymerase, extracytoplasmic E) encodes the sigma factor ''sigma-24'' (σ24, sigma E, or RpoE), a protein in ''Escherichia coli'' and other species of bacteria. Depending on the bacterial species, this gene may be referred to as ''sigE''. RpoE appears to be necessary for the exocytoplasmic stress response. ''E. coli'' mutants without ''rpoE'' cannot grow at high temperatures (that is, above 42 degrees C) and show growth defects at lower temperatures, though this may be due to compensatory mutations. In some bacterial species, such as ''Clostridium botulinum'', this sigma factor may be necessary for sporulation In biology, a spore is a unit of sexual (in fungi) or asexual reproduction that may be adapted for dispersal and for survival, often for extended periods of time, in unfavourable conditions. Spores form part of the life cycles of many plant .... The stress response regulation activities of RpoE are modulated by the Hfq protein in ''E. coli''. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RpoF
The gene ''rpoF'' (RNA polymerase, flagellum F) encodes the sigma factor ''sigma-28'' (σ28, or RpoF), a protein in ''Escherichia coli'' and other species of bacteria. Depending on the bacterial species, this gene may be referred to as ''sigD'' or ''fliA''. The protein encoded by this gene has been found to be necessary for flagellum A flagellum (; : flagella) (Latin for 'whip' or 'scourge') is a hair-like appendage that protrudes from certain plant and animal sperm cells, from fungal spores ( zoospores), and from a wide range of microorganisms to provide motility. Many pr ... formation. References Escherichia coli genes {{gene-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stress Response
The fight-or-flight or the fight-flight-freeze-or-fawn (also called hyperarousal or the acute stress response) is a physiological reaction that occurs in response to a perceived harmful event, attack, or threat to survival. It was first described by Walter Bradford Cannon in 1915. His theory states that animals react to threats with a general discharge of the sympathetic nervous system, preparing the animal for fighting or fleeing. More specifically, the adrenal medulla produces a hormonal cascade that results in the secretion of catecholamines, especially norepinephrine and epinephrine. The hormones estrogen, testosterone, and cortisol, as well as the neurotransmitters dopamine and serotonin, also affect how organisms react to stress. The hormone osteocalcin might also play a part. This response is recognised as the first stage of the general adaptation syndrome that regulates stress responses among vertebrates and other organisms. Name Originally understood as the "fig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RpoS
The gene ''rpoS'' (RNA polymerase, sigma S, also called katF) encodes the sigma factor ''sigma-38'' (σ38, or RpoS), a 37.8 kD protein in ''Escherichia coli''. Sigma factors are proteins that regulate transcription (genetics), transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. ''rpoS'' is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The ''rpoS'' gene most likely originated in the gammaproteobacte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Negative Feedback
Negative feedback (or balancing feedback) occurs when some function (Mathematics), function of the output of a system, process, or mechanism is feedback, fed back in a manner that tends to reduce the fluctuations in the output, whether caused by changes in the input or by other disturbances. Whereas positive feedback tends to instability via exponential growth, oscillation or chaos theory, chaotic behavior, negative feedback generally promotes stability. Negative feedback tends to promote a settling to List of types of equilibrium, equilibrium, and reduces the effects of perturbations. Negative feedback loops in which just the right amount of correction is applied with optimum timing, can be very stable, accurate, and responsive. Negative feedback is widely used in Mechanical engineering, mechanical and electronic engineering, and it is observed in many other fields including biology, chemistry and economics. General negative feedback systems are studied in Control engin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (biology)
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transcribed into RNA molecules called non-coding RNAs (ncRNAs). Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a Complementarity (molecular biology), complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, Antiparallel (biochemistry), antiparallel RNA strand called a primary transcript. In virology, the term transcription is used when referring to mRNA synthesis from a viral RNA molecule. The genome of many Orthornavirae, RNA viruses is composed of Sense (molecular biology), negative-sense RNA which acts as a template for positive sense viral messenger RNA - a necessary step in the synthesis of viral proteins needed for viral replication. This process ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulated Intramembrane Proteolysis
Intramembrane proteases (IMPs), also known as intramembrane-cleaving proteases (I-CLiPs), are enzymes that have the property of cleaving transmembrane domains of integral membrane proteins. All known intramembrane proteases are themselves integral membrane proteins with multiple transmembrane domains, and they have their active sites buried within the lipid bilayer of cellular membranes. Intramembrane proteases are responsible for proteolytic cleavage in the cell signaling process known as regulated intramembrane proteolysis (RIP). Intramembrane proteases are not evolutionarily related to classical soluble proteases, having evolved their catalytic sites by convergent evolution. Although only recently discovered, intramembrane proteases are of significant research interest because of their major biological functions and their relevance to human disease. Classification There are four groups of intramembrane proteases, distinguished by their catalytic mechanism: * Metalloproteases ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dephosphorylation
In biochemistry, dephosphorylation is the removal of a phosphate () group from an organic compound by hydrolysis. It is a reversible post-translational modification. Dephosphorylation and its counterpart, phosphorylation, activate and deactivate enzymes by detaching or attaching phosphoric esters and anhydrides. A notable occurrence of dephosphorylation is the conversion of Adenosine triphosphate, ATP to Adenosine diphosphate, ADP and inorganic phosphate. Dephosphorylation employs a type of hydrolytic enzyme, or hydrolase, which cleaves ester bonds. The prominent hydrolase subclass used in dephosphorylation is phosphatase, which removes phosphate groups by hydrolysing phosphoric acid monoesters into a phosphate ion and a molecule with a free hydroxyl (–OH) group. The reversible phosphorylation-dephosphorylation reaction occurs in every physiological process, making proper function of protein phosphatases necessary for organism viability. Because protein dephosphorylation is a k ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |