|
Actinomycete
The Actinomycetales is an order of Actinomycetota. A member of the order is often called an actinomycete. Actinomycetales are generally gram-positive and anaerobic and have mycelia in a filamentous and branching growth pattern. Some actinomycetes can form rod- or coccoid-shaped forms, while others can form spores on aerial hyphae. Actinomycetales bacteria can be infected by bacteriophages, which are called actinophages. Actinomycetales can range from harmless bacteria to pathogens with resistance to antibiotics. Reproduction Actinomycetales have 2 main forms of reproduction: spore formation and hyphae fragmentation. During reproduction, Actinomycetales can form conidiophores, sporangiospores, and oidiospores. In reproducing through hyphae fragmentation, the hyphae formed by Actinomycetales can be a fifth to half the size of fungal hyphae, and bear long spore chains. Presence and associations Actinomycetales can be found mostly in soil and decaying organic matter, as well as in l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Actinomycetota
The Actinomycetota (or Actinobacteria) are a diverse phylum of Gram-positive bacteria with high GC content. They can be terrestrial or aquatic. They are of great importance to land flora because of their contributions to soil systems. In soil they help to decompose the organic matter of dead organisms so the molecules can be taken up anew by plants. While this role is also played by fungi, Actinomycetota are much smaller and likely do not occupy the same ecological niche. In this role the colonies often grow extensive mycelia, as fungi do, and the name of an important order of the phylum, Actinomycetales (the actinomycetes), reflects that they were long believed to be fungi. Some soil actinomycetota (such as '' Frankia'') live symbiotically with the plants whose roots pervade the soil, fixing nitrogen for the plants in exchange for access to some of the plant's saccharides. Other species, such as many members of the genus ''Mycobacterium'', are important pathogens. Beyond th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Actinomyces
''Actinomyces'' is a genus of the Actinomycetia class of bacteria. They all are Gram-positive and facultatively anaerobic, growing best under anaerobic conditions. ''Actinomyces'' species may form endospores, and while individual bacteria are rod-shaped, ''Actinomyces'' colonies form fungus-like branched networks of hyphae. The aspect of these colonies initially led to the incorrect assumption that the organism was a fungus and to the name ''Actinomyces'', "ray fungus" (from Greek , ray or beam, and , fungus). ''Actinomyces'' species are ubiquitous, occurring in soil and in the microbiota of animals, including the human microbiota. They are known for the important role they play in soil ecology; they produce a number of enzymes that help degrade organic plant material, lignin, and chitin. Thus, their presence is important in the formation of compost. Certain species are commensal in the skin flora, oral flora, gut flora, and vaginal flora of humans and livestock. They are also ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
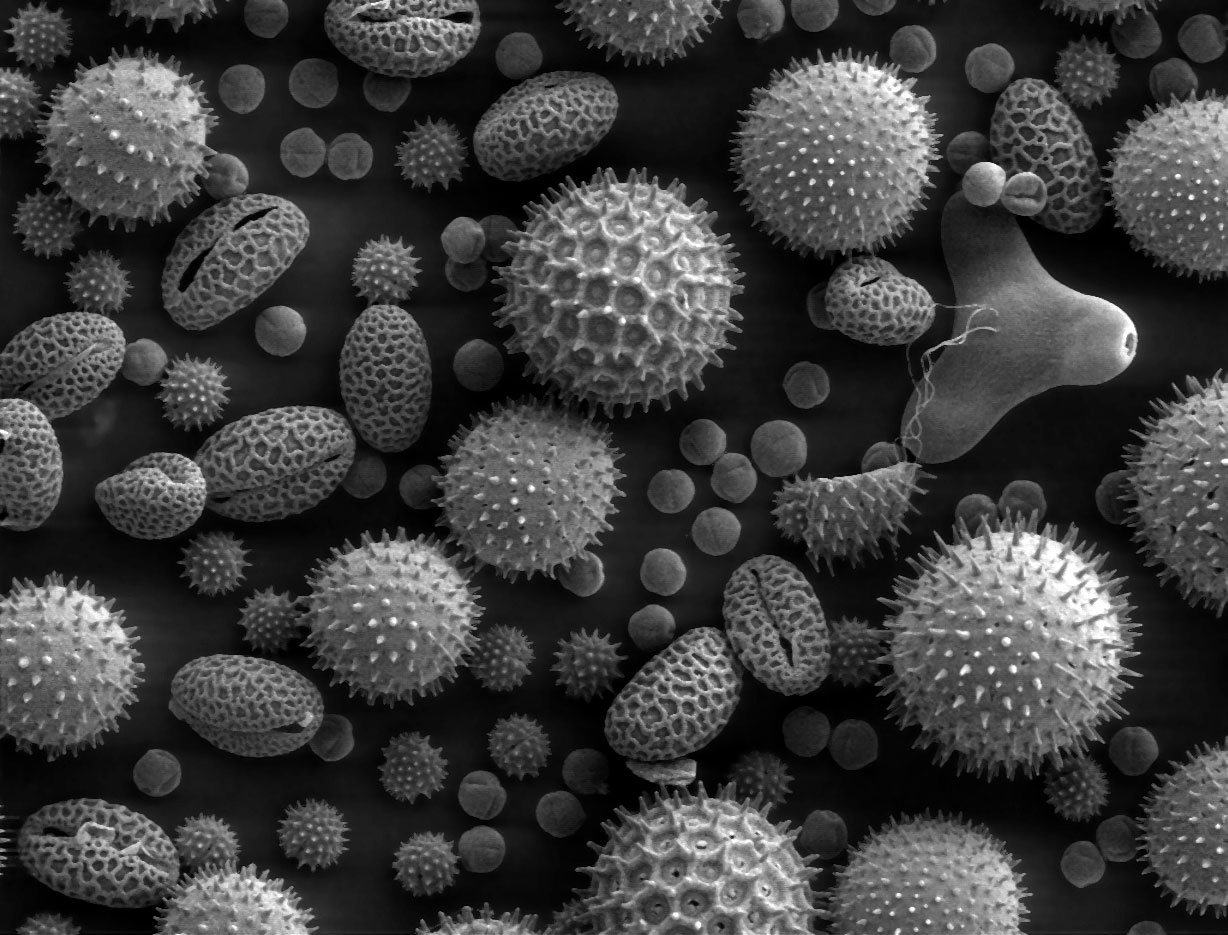
Scanning Electron Micrograph
A scanning electron microscope (SEM) is a type of electron microscope that produces images of a sample by scanning the surface with a focused beam of electrons. The electrons interact with atoms in the sample, producing various signals that contain information about the surface topography and composition. The electron beam is scanned in a raster scan pattern, and the position of the beam is combined with the Intensity (physics), intensity of the detected signal to produce an image. In the most common SEM mode, secondary electrons emitted by atoms excited by the electron beam are detected using a secondary electron detector (Everhart–Thornley detector). The number of secondary electrons that can be detected, and thus the signal intensity, depends, among other things, on specimen topography. Some SEMs can achieve resolutions better than 1 Nanometre, nanometer. Specimens are observed in high vacuum in a Convention (norm), conventional SEM, or in low vacuum or wet conditions in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Kytococcaceae
''Kytococcus'' is a genus of bacteria in the phylum Actinomycetota The Actinomycetota (or Actinobacteria) are a diverse phylum of Gram-positive bacteria with high GC content. They can be terrestrial or aquatic. They are of great importance to land flora because of their contributions to soil systems. In soil t .... References Micrococcales Bacteria genera {{Actinobacteria-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Kineosporiaceae
The Kineosporiaceae is a family of Gram positive bacteria. Phylogeny The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI). Notes See also * List of bacterial orders * List of bacteria genera This article lists the genera of the bacteria Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, ... References Actinomycetia {{actinobacteria-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microbacteriaceae
Microbacteriaceae is a family of bacteria of the order Actinomycetales. They are Gram-positive soil organisms. Genera The family ''Microbacteriaceae'' comprises the following genera: * ''Agreia'' Evtushenko ''et al''. 2001 * ''Agrococcus'' Groth ''et al''. 1996 * ''Agromyces'' Gledhill and Casida 1969 (Approved Lists 1980) * ''Allohumibacter'' Kim ''et al''. 2016 * ''Alpinimonas'' Schumann ''et al''. 2012 * ''Amnibacterium'' Kim and Lee 2011 * ''Arenivirga'' Hamada ''et al''. 2017 * ''Aurantimicrobium'' Nakai ''et al''. 2015 * ''Canibacter'' Aravena-Román ''et al''. 2014 * ''Clavibacter'' Davis ''et al''. 1984 * ''Cnuibacter'' Zhou ''et al''. 2016 * ''Compostimonas'' Kim ''et al''. 2012 * ''Conyzicola'' Kim ''et al''. 2014 * "''Crocebacterium''" Rogers & Doran-Peterson 2006 * ''Cryobacterium'' Suzuki ''et al''. 1997 * "''Cryocola''" Gavrish ''et al''. 2003 * ''Curtobacterium'' Yamada and Komagata 1972 (Approved Lists 1980) * ''Diaminobutyricibacter'' Kim ''et al''. 2014 * ''D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microbacteriales
The Micrococcales are an order of bacteria in the phylum Actinomycetota. Phylogeny The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI) Notes See also * Bacterial taxonomy * List of bacterial orders * List of bacteria genera This article lists the genera of the bacteria Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, ... References Actinomycetia Bacteria orders {{Actinobacteria-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Taxonomy Database
The Genome Taxonomy Database (GTDB) is an online database that maintains information on a proposed nomenclature of prokaryotes, following a phylogenomic approach based on a set of conserved single-copy proteins. In addition to resolving paraphyletic groups, this method also reassigns taxonomic ranks algorithmically, updating names in both cases. Information for archaea was added in 2020, along with a species classification based on average nucleotide identity. Each update incorporates new genomes as well as automated and manual curation of the taxonomy. An open-source tool called GTDB-Tk is available to classify draft genomes into the GTDB hierarchy. The GTDB system, via GTDB-Tk, has been used to catalogue not-yet-named bacteria in the human gut microbiome and other metagenomic sources. The GTDB is incorporated into the '' Bergey's Manual of Systematics of Archaea and Bacteria'' in 2019 as its phylogenomic resource. Methodology The genomes used to construct the phyloge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
The All-Species Living Tree Project
The All-Species Living Tree' Project is a collaboration between various academic groups/institutes, such as ARB, SILVA rRNA database project, and LPSN, with the aim of assembling a database of 16S rRNA sequences of all validly published species of ''Bacteria'' and ''Archaea''. At one stage, 23S sequences were also collected, but this has since stopped. Currently there are over 10,950 species in the aligned dataset and several more are being added either as new species are discovered or species that are not represented in the database are sequenced. Initially the latter group consisted of 7% of species. Similar (and more recent) projects include the Genomic Encyclopedia of Bacteria and Archaea (GEBA), which focused on whole genome sequencing of bacteria and archaea. Tree The tree was created by maximum likelihood analysis without bootstrap: consequently accuracy is traded off for size and many phylum level clades are not correctly resolved (such as the Firmicutes). (Eukaryote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
National Center For Biotechnology Information
The National Center for Biotechnology Information (NCBI) is part of the National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). It is approved and funded by the government of the United States. The NCBI is located in Bethesda, Maryland, and was founded in 1988 through legislation sponsored by US Congressman Claude Pepper. The NCBI houses a series of databases relevant to biotechnology and biomedicine and is an important resource for bioinformatics tools and services. Major databases include GenBank for DNA sequences and PubMed, a bibliographic database for biomedical literature. Other databases include the NCBI Epigenomics database. All these databases are available online through the Entrez search engine. NCBI was directed by David Lipman, one of the original authors of the BLAST sequence alignment program and a widely respected figure in bioinformatics. GenBank NCBI had responsibility for making available the GenBank DNA seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |