|
8-hydroxydeoxyguanosine
8-Oxo-2'-deoxyguanosine (8-oxo-dG) is an oxidized derivative of deoxyguanosine. 8-Oxo-dG is one of the major products of DNA oxidation. Concentrations of 8-oxo-dG within a cell are a measurement of oxidative stress. In DNA Steady-state levels of DNA damages represent the balance between formation and repair. Swenberg et al. measured average frequencies of steady state endogenous DNA damages in mammalian cells. The most frequent oxidative DNA damage normally present in DNA is 8-oxo-dG, occurring at an average frequency of 2,400 per cell. When 8-oxo-dG is induced by a DNA damaging agent it is rapidly repaired. For example, 8-oxo-dG was increased 10-fold in the livers of mice subjected to ionizing radiation, but the excess 8-oxo-dG was rapidly removed with a half-life of 11 minutes. As reviewed by Valavanidis et al. increased levels of 8-oxo-dG in a tissue can serve as a biomarker of oxidative stress. They also noted that increased levels of 8-oxo-dG are frequently found du ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Oxidation
DNA oxidation is the process of oxidative damage of Deoxyribonucleic Acid, deoxyribonucleic acid. As described in detail by Burrows et al., 8-oxo-2'-deoxyguanosine (8-oxo-dG) is the most common oxidative lesion observed in duplex DNA because guanine has a lower one-electron Redox, reduction potential than the other nucleosides in DNA. The one electron reduction potentials of the nucleosides (in volts versus Standard hydrogen electrode#Relationship between the normal hydrogen electrode (NHE) and the standard hydrogen electrode (SHE), NHE) are guanine 1.29, adenine 1.42, cytosine 1.6 and thymine 1.7. About 1 in 40,000 guanines in the genome are present as 8-oxo-dG under normal conditions. This means that >30,000 8-oxo-dGs may exist at any given time in the genome of a human cell. Another product of DNA oxidation is 8-oxo-dA. 8-oxo-dA occurs at about 1/10 the frequency of 8-oxo-dG. The reduction potential of guanine may be reduced by as much as 50%, depending on the particular ne ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Damage Theory Of Aging
The DNA damage theory of aging proposes that aging is a consequence of unrepaired accumulation of DNA damage (naturally occurring), naturally occurring DNA damage. Damage in this context is a DNA alteration that has an abnormal structure. Although both mitochondrion, mitochondrial and Cell nucleus, nuclear DNA damage can contribute to aging, nuclear DNA is the main subject of this analysis. Nuclear DNA damage can contribute to aging either indirectly (by increasing apoptosis or Hayflick limit, cellular senescence) or directly (by increasing cell dysfunction). Several review articles have shown that deficient DNA repair, allowing greater accumulation of DNA damage, causes premature aging; and that increased DNA repair facilitates greater longevity, e.g. Mouse models of nucleotide-excision–repair syndromes reveal a striking correlation between the degree to which specific DNA repair pathways are compromised and the severity of accelerated aging, strongly suggesting a causal relation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
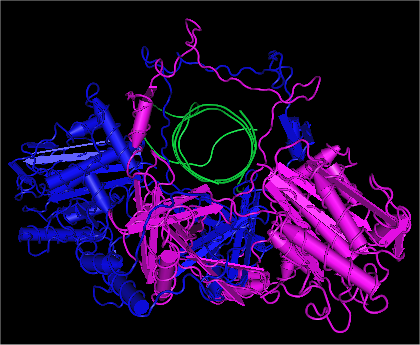
OGG1
8-Oxoguanine glycosylase, also known as OGG1, is a DNA glycosylase enzyme that, in humans, is encoded by the ''OGG1'' gene. It is involved in base excision repair. It is found in bacterial, archaeal and eukaryotic species. Function OGG1 is the primary enzyme responsible for the excision of 8-oxoguanine (8-oxoG), a mutagenic base byproduct that occurs as a result of exposure to reactive oxygen species (ROS). OGG1 is a bifunctional glycosylase, as it is able to both cleave the glycosidic bond of the mutagenic lesion and cause a strand break in the DNA backbone. Alternative splicing of the C-terminal region of this gene classifies splice variants into two major groups, type 1 and type 2, depending on the last exon of the sequence. Type 1 alternative splice variants end with exon 7 and type 2 end with exon 8. One set of spliced forms are designated 1a, 1b, 2a to 2e. All variants have the N-terminal region in common. Many alternative splice variants for this gene have been de ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in Cell (biology), cells, by internal metabolism, metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for Transcription (biology), transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Damage
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in cells, by internal metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair processes fail, including apoptosis, irreparable DNA damage may occur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
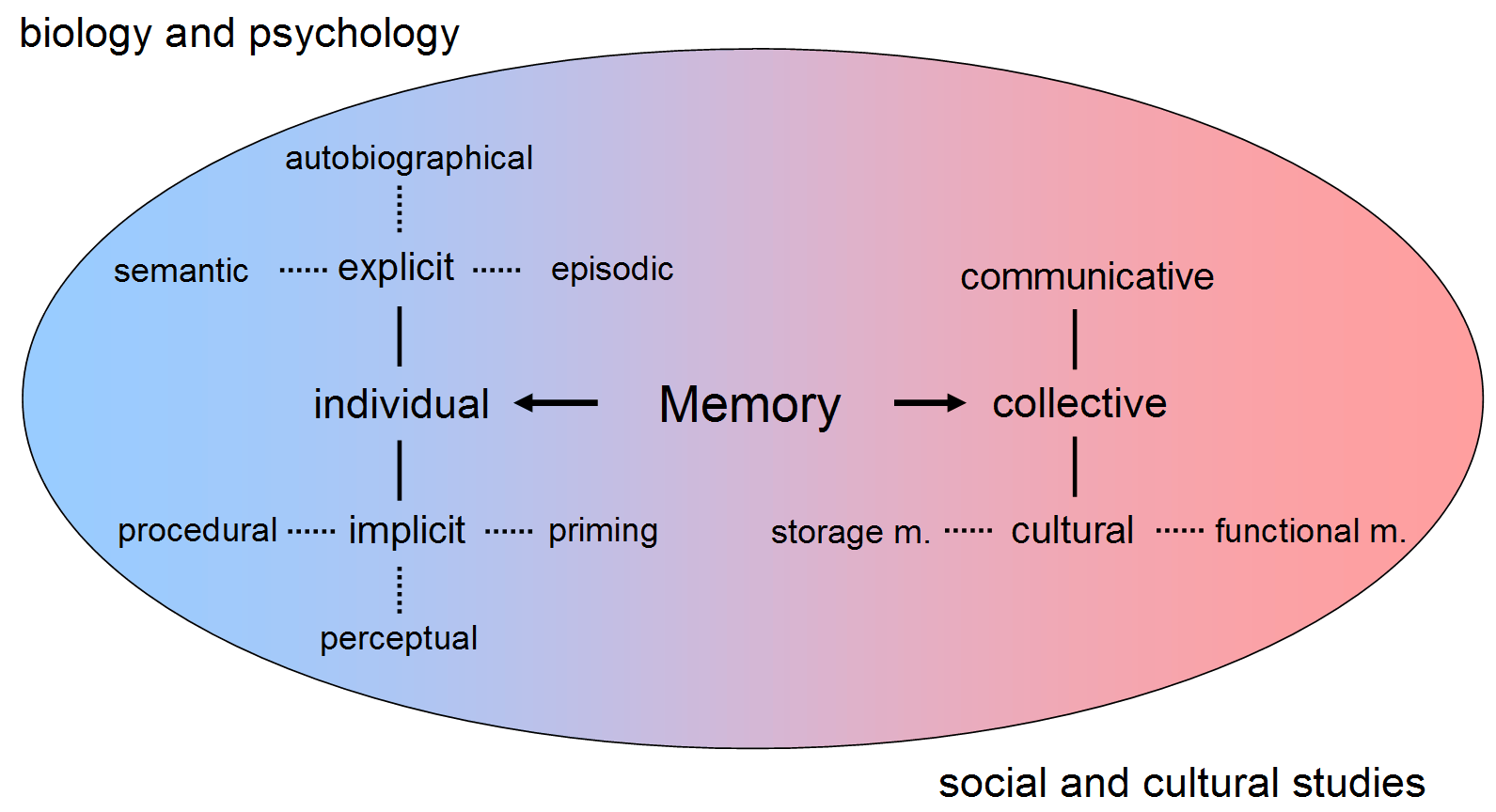
Memory
Memory is the faculty of the mind by which data or information is encoded, stored, and retrieved when needed. It is the retention of information over time for the purpose of influencing future action. If past events could not be remembered, it would be impossible for language, relationships, or personal identity to develop. Memory loss is usually described as forgetfulness or amnesia. Memory is often understood as an informational processing system with explicit and implicit functioning that is made up of a sensory processor, short-term (or working) memory, and long-term memory. This can be related to the neuron. The sensory processor allows information from the outside world to be sensed in the form of chemical and physical stimuli and attended to various levels of focus and intent. Working memory serves as an encoding and retrieval processor. Information in the form of stimuli is encoded in accordance with explicit or implicit functions by the working memory p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
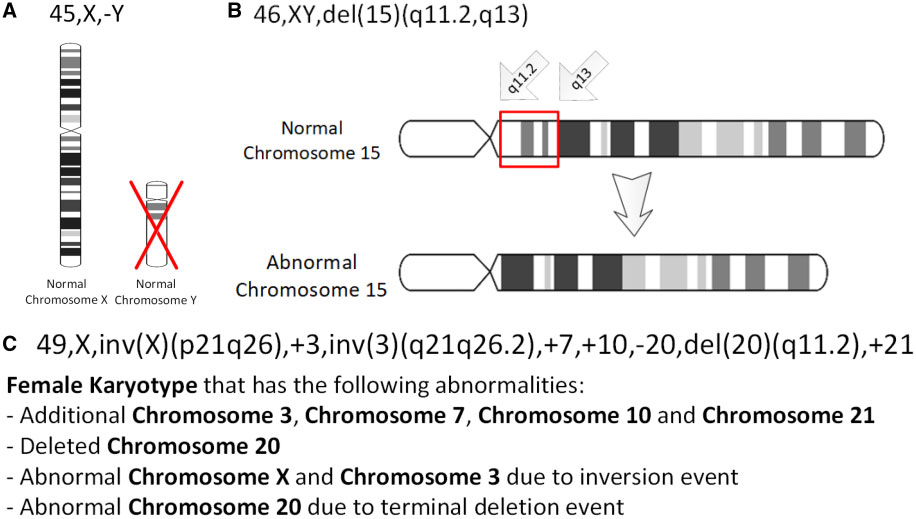
Deletion (genetics)
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur, which result in the deletion of a part of the chromosome. The breaks can be induced by heat, viruses, radiation, or chemical reactions. When a chromosome breaks, if a part of it is deleted or lost, the missing piece of chromosome is referred to as a deletion or a deficiency. For synapsis to occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into a deletion or compensation loop. The smallest single base deletion mutations occur by a single base flipping in the template DNA, followed by te ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transversion
Transversion, in molecular biology, refers to a point mutation in DNA in which a single (two ring) purine ( A or G) is changed for a (one ring) pyrimidine ( T or C), or vice versa. A transversion can be spontaneous, or it can be caused by ionizing radiation or alkylating agents. It can only be reversed by a spontaneous reversion. Ratio of transitions to transversions Although there are two possible transversions but only one possible transition per base, transition mutations are more likely than transversions because substituting a single ring structure for another single ring structure is more likely than substituting a double ring for a single ring. Also, transitions are less likely to result in amino acid substitutions (due to wobble base pair), and are therefore more likely to persist as "silent substitutions" in populations as single nucleotide polymorphisms (SNPs). A transversion usually has a more pronounced effect than a transition because the second and third nucle ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer in adults and is currently the most common cause of death in people with cirrhosis. HCC is the third leading cause of cancer-related deaths worldwide. HCC most commonly occurs in those with chronic liver disease especially those with cirrhosis or fibrosis, which occur in the setting of chronic liver injury and inflammation. HCC is rare in those without chronic liver disease. Chronic liver diseases which greatly increase the risk of HCC include hepatitis infection such as (hepatitis B, hepatitis C, C or hepatitis D, D), non-alcoholic steatohepatitis (NASH), alcoholic liver disease, or exposure to toxins such as aflatoxin, or pyrrolizidine alkaloids. Certain diseases, such as HFE hereditary haemochromatosis, hemochromatosis and alpha 1-antitrypsin deficiency, markedly increase the risk of developing HCC. The five-year survival in those with HCC is 18%. As with any cancer, the treatment and prognosis of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thymidine Kinase
Thymidine kinase is an enzyme, a phosphotransferase (a kinase): 2'-deoxythymidine kinase, ATP-thymidine 5'-phosphotransferase, EC 2.7.1.21. It can be found in most living cells. It is present in two forms in mammalian cells, TK1 and TK2. Certain viruses also have genetic information for expression of viral thymidine kinases. Thymidine kinase catalyzes the reaction: :Thd + ATP → TMP + ADP where Thd is (deoxy)thymidine, ATP is adenosine triphosphate, TMP is (deoxy) thymidine monophosphate and ADP is adenosine diphosphate. Thymidine kinases have a key function in the synthesis of DNA and therefore in cell division, as they are part of the unique reaction chain to introduce thymidine into the DNA. Thymidine is present in the body fluids as a result of degradation of DNA from food and from dead cells. Thymidine kinase is required for the action of many antiviral drugs. It is used to select hybridoma cell lines in production of monoclonal antibodies. In clinical chemistry it is u ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Base Excision Repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair pathway repairs bulky helix-distorting lesions. BER is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP sites. These are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch (where a single nucleotide is replaced) or long-patch BER (where 2–10 new nucleotides are synthesized). Lesions processed by BER Single bases in DNA can be chemically damaged by a variety of mechanisms, the most common ones being deamination, oxidation, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cancer Epigenetics
Cancer epigenetics is the study of epigenetics, epigenetic modifications to the DNA of cancer cells that do not involve a change in the nucleotide sequence, but instead involve a change in the way the genetic code is expressed. Epigenetic mechanisms are necessary to maintain normal sequences of tissue specific gene expression and are crucial for normal development. They may be just as important, if not even more important, than mutation, genetic mutations in a cell's transformation to cancer. The disturbance of epigenetic processes in cancers, can lead to a loss of gene expression, expression of genes that occurs about 10 times more frequently by transcription silencing (caused by epigenetic promoter hypermethylation of CpG site#Methylation, silencing, cancer, and aging, CpG islands) than by mutations. As Vogelstein et al. points out, in a colorectal cancer there are usually about 3 to 6 driver mutations and 33 to 66 Genetic hitchhiking, hitchhiker or passenger mutations. However ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |