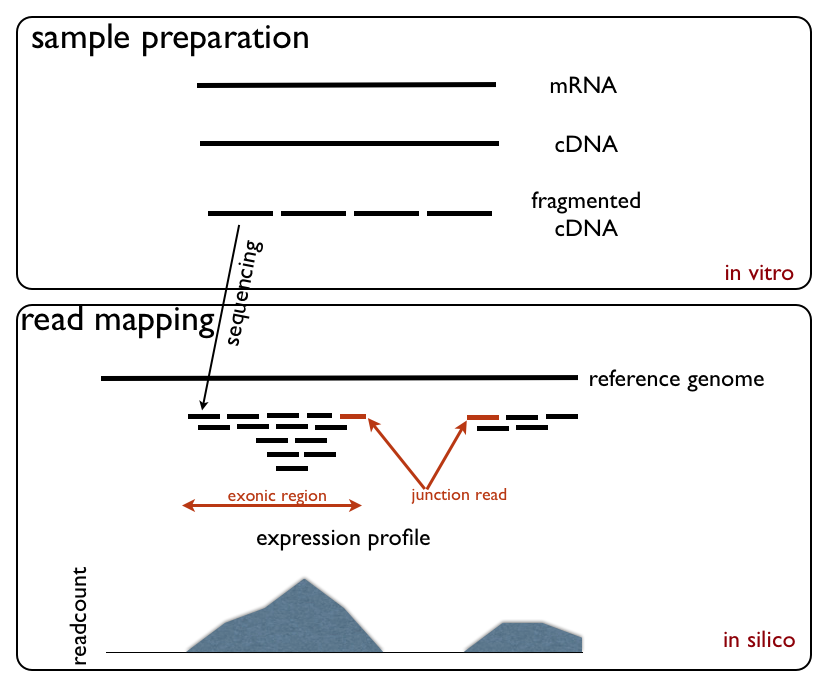
|
3' MRNA-seq
3' mRNA-seq is a quantitative, genome-wide transcriptomic technique based on the barcoding of the 3' untranslated region (UTR) of mRNA molecules. Unlike standard bulk RNA-seq, where short sequencing reads are generated along the entire length of mRNA transcripts, only the 3' end of polyadenylated RNAs are sequenced in 3' mRNA-seq. This approach results in a need for fewer reads to quantify the expression of a gene and reduces the sequencing depth required per sample while providing robust and reliable transcriptome-wide read-outs of gene expression levels comparable to full-length RNA-seq methods. Sample barcoding and the reduced per-sample sequencing depth also allow higher levels of sample multiplexing per experiment and lower the cost of transcriptome sequencing compared to full-length RNA-seq methods. These factors are crucial for large-scale, ultra-high-throughput gene expression studies or studies assessing differential gene expression between different experimental cond ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcriptomics Technologies
Transcriptomics technologies are the techniques used to study an organism's transcriptome, the sum of all of its RNA transcripts. The information content of an organism is recorded in the DNA of its genome and expressed through transcription. Here, mRNA serves as a transient intermediary molecule in the information network, whilst non-coding RNAs perform additional diverse functions. A transcriptome captures a snapshot in time of the total transcripts present in a cell. Transcriptomics technologies provide a broad account of which cellular processes are active and which are dormant. A major challenge in molecular biology is to understand how a single genome gives rise to a variety of cells. Another is how gene expression is regulated. The first attempts to study whole transcriptomes began in the early 1990s. Subsequent technological advances since the late 1990s have repeatedly transformed the field and made transcriptomics a widespread discipline in biological sciences. There a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Three Prime Untranslated Region
In molecular genetics, the three prime untranslated region (3′-UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codon. The 3′-UTR often contains regulatory regions that post-transcriptionally influence gene expression. During gene expression, an mRNA molecule is transcribed from the DNA sequence and is later translated into a protein. Several regions of the mRNA molecule are not translated into a protein including the 5' cap, 5' untranslated region, 3′ untranslated region and poly(A) tail. Regulatory regions within the 3′-untranslated region can influence polyadenylation, translation efficiency, localization, and stability of the mRNA. The 3′-UTR contains binding sites for both regulatory proteins and microRNAs (miRNAs). By binding to specific sites within the 3′-UTR, miRNAs can decrease gene expression of various mRNAs by either inhibiting translation or directly causing degradation of the transcript. The 3′-U ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and the ribosome creates the protein utilizing amino acids carried by transfer RNA (tRNA). This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, genetic inf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also known as transcriptome. RNA-Seq facilitates the ability to look at alternative gene spliced transcripts, post-transcriptional modifications, gene fusion, mutations/ SNPs and changes in gene expression over time, or differences in gene expression in different groups or treatments. In addition to mRNA transcripts, RNA-Seq can look at different populations of RNA to include total RNA, small RNA, such as miRNA, tRNA, and ribosomal profiling. RNA-Seq can also be used to determine exon/intron boundaries and verify or amend previously annotated 5' and 3' gene boundaries. Recent advances in RNA-Seq include single cell sequencing, bulk RNA sequencing, 3' mRNA-sequencing, in situ sequencing of fixed tissue, and native RNA molecule sequencin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Read (biology)
In DNA sequencing, a read is an inferred sequence of base pairs (or base pair probabilities) corresponding to all or part of a single DNA fragment. A typical sequencing experiment involves DNA fragmentation, fragmentation of the genome into millions of molecules, which are size-selected and ligation (molecular biology), ligated to adapter (genetics), adapters. The set of fragments is referred to as a sequencing library, which is sequenced to produce a set of reads. Read length Sequencing technologies vary in the length of reads produced. Reads of length 20-40 base pairs (bp) are referred to as ultra-short. Typical sequencers produce read lengths in the range of 100-500 bp. However, Pacific Biosciences platforms produce read lengths of approximately 1500 bp. Read length is a factor which can affect the results of biological studies. For example, longer read lengths improve the resolution of ''de novo'' genome assembly and detection of structural variants. It is estimated that read ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression. The process of polyadenylation begins as the transcription of a gene terminates. The 3′-most segment of the newly made pre-mRNA is first cleaved off by a set of proteins; these proteins then synthesize the poly(A) tail at the RNA's 3′ end. In some genes these proteins add a poly(A) tail at one of several possible sites. Therefore, polyadenylation can produce more than one transcript from a single gene (alternative polyadenylation), similar to alternative splicing. The poly(A) tail is important for the nuclea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequencing Depth
In genetics, coverage is one of several measures of the depth or completeness of DNA sequencing, and is more specifically expressed in any of the following terms: *Sequence coverage (or depth) is the number of unique reads that include a given nucleotide in the reconstructed sequence. Deep sequencing refers to the general concept of aiming for high number of unique reads of each region of a sequence. *Physical coverage, the cumulative length of reads or read pairs expressed as a multiple of genome size. *Genomic coverage, the percentage of all base pairs or loci of the genome covered by sequencing. Sequence coverage Rationale Even though the sequencing accuracy for each individual nucleotide is very high, the very large number of nucleotides in the genome means that if an individual genome is only sequenced once, there will be a significant number of sequencing errors. Furthermore, many positions in a genome contain rare single-nucleotide polymorphisms (SNPs). Hence to distingu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BRB-seq
Bulk RNA barcoding and sequencing (BRB-seq) is an ultra-high-throughput bulk 3' mRNA-seq technology that uses early-stage sample barcoding and Unique molecular identifier, unique molecular identifiers (UMIs) to allow the pooling of up to 384 samples in one tube early in the sequencing library preparation workflow. The transcriptomic technology is compatible with both Illumina, Inc., Illumina and MGI (company), MGI short-read sequencing instruments. In standard RNA-Seq, RNA-seq, a sequencing library must be prepared for each RNA sample individually. In contrast, in BRB-seq, all samples are pooled early in the workflow for simultaneous processing to reduce the cost and hands-on time associated with the library preparation stage As BRB-seq is a 3' mRNA-seq technique, Short-read sequencing, short reads are generated only for the 3' region of polyadenylated mRNA molecules instead of the full length of transcripts like in standard RNA-Seq, RNA-seq. This means that BRB-seq requires a fa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Unique Molecular Identifier
Unique molecular identifiers (UMIs), or molecular barcodes (MBC) are short sequences or molecular "tags" added to DNA fragments in some next generation sequencing library preparation protocols to identify the input DNA molecule. These tags are added before PCR amplification, and can be used to reduce errors and quantitative bias introduced by the amplification. Applications include analysis of unique cDNAs to avoid PCR biases in iCLIP, variant calling in ctDNA, gene expression in single-cell RNA-seq (scRNA-seq) and haplotyping via linked reads. See also * Batch effect * Multiplex (assay) *BRB-seq Bulk RNA barcoding and sequencing (BRB-seq) is an ultra-high-throughput bulk 3' mRNA-seq technology that uses early-stage sample barcoding and Unique molecular identifier, unique molecular identifiers (UMIs) to allow the pooling of up to 384 samp ... References DNA sequencing {{genetics-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research and forensic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-cell Transcriptomics
Single-cell transcriptomics examines the gene expression level of individual Cell (biology), cells in a given population by simultaneously measuring the RNA concentration (conventionally only messenger RNA (mRNA)) of hundreds to thousands of genes. Single-cell transcriptomics makes it possible to unravel Heterogenous, heterogeneous cell populations, reconstruct cellular developmental pathways, and model transcriptional dynamics — all previously masked in bulk RNA sequencing. Background The development of high-throughput RNA sequencing (RNA-seq) and microarrays has made gene expression analysis a routine. RNA analysis was previously limited to tracing individual Transcription (biology), transcripts by Northern blots or quantitative PCR. Higher throughput and speed allow researchers to frequently characterize the expression profiles of populations of thousands of cells. The data from bulk assays has led to identifying genes differentially expressed in distinct cell populations, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcriptase
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. The process does not violate the flows of genetic information as described by the classical central dogma, but rather expands it to include transfers of information from RNA to DNA. Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host genome, from which new RNA copies can be made via host-cell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |