Molecular evolution on:
[Wikipedia]
[Google]
[Amazon]
Molecular evolution is the process of change in the sequence composition of cellular
 Mutations are permanent, transmissible changes to the
Mutations are permanent, transmissible changes to the
 Recombination is a process that results in genetic exchange between chromosomes or chromosomal regions. Recombination counteracts physical linkage between adjacent genes, thereby reducing genetic hitchhiking. The resulting independent inheritance of genes results in more efficient selection, meaning that regions with higher recombination will harbor fewer detrimental mutations, more selectively favored variants, and fewer errors in replication and repair. Recombination can also generate particular types of mutations if chromosomes are misaligned.
Recombination is a process that results in genetic exchange between chromosomes or chromosomal regions. Recombination counteracts physical linkage between adjacent genes, thereby reducing genetic hitchhiking. The resulting independent inheritance of genes results in more efficient selection, meaning that regions with higher recombination will harbor fewer detrimental mutations, more selectively favored variants, and fewer errors in replication and repair. Recombination can also generate particular types of mutations if chromosomes are misaligned.
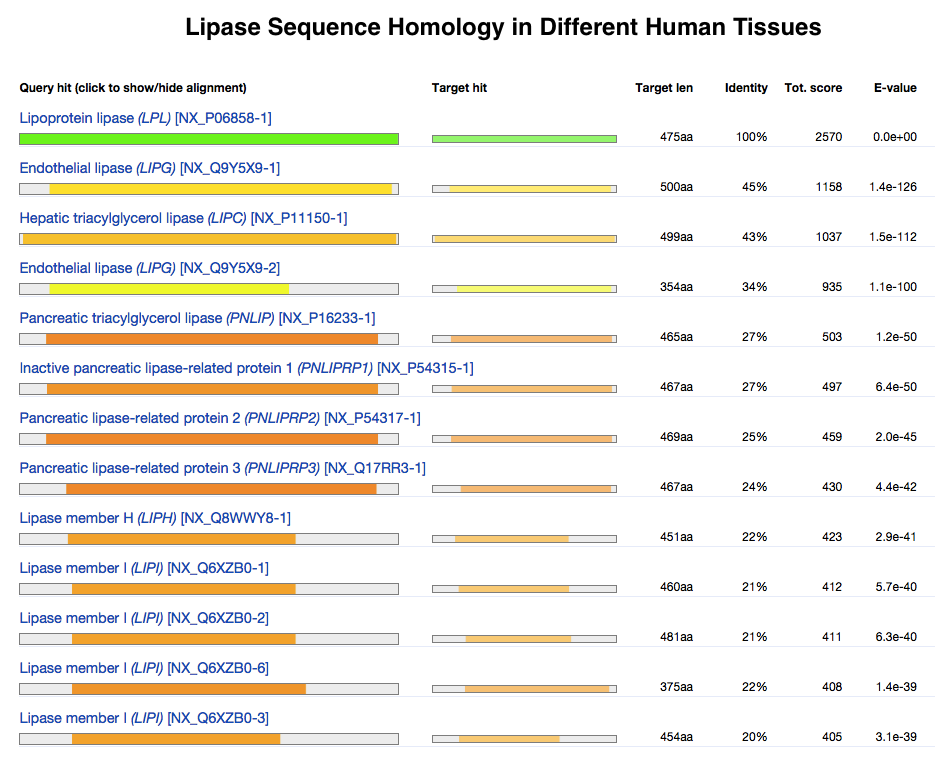 Evolution of proteins is studied by Sequence alignment, comparing the sequences and Protein structure, structures of proteins from many organisms. Similar sequences/structures indicating that the proteins diverged from a common origin; these proteins are ''Homology (biology), homologous''. Phylogenetics, Phylogenetic analysis of proteins has revealed how proteins evolve and change their structure and function over time.
Evolutionary rate. Using the amino acid sequences of hemoglobin and cytochrome c from multiple species, scientists were able to derive estimations of protein evolution rates. Each protein has its own rate, and that rate is relatively constant across phylogenies (i.e., hemoglobin does not evolve at the same rate as cytochrome c, but hemoglobins from humans, mice, etc. do have comparable rates of evolution). Not all regions within a protein mutate at the same rate; functionally important areas mutate more slowly and amino acid substitutions involving similar amino acids occurs more often than dissimilar substitutions. Overall, the level of polymorphisms in proteins seems to be fairly constant. Several species (including humans, fruit flies, and mice) have similar levels of protein polymorphism.
Functional evolution. Numerous enzymes and other proteins have been shown to change their function over the course of evolution. For example, ribonucleotide reductase (RNR) is known from thousands of organisms and has evolved a multitude of structural and functional variants. Class I RNRs use a ferritin subunit and differ by the metal they use as cofactors. In class II RNRs, the thiyl radical is generated using an adenosylcobalamin cofactor and these enzymes do not require additional subunits (as opposed to class I which do). In class III RNRs, the thiyl radical is generated using S-Adenosyl methionine, S-adenosylmethionine bound to a [
Evolution of proteins is studied by Sequence alignment, comparing the sequences and Protein structure, structures of proteins from many organisms. Similar sequences/structures indicating that the proteins diverged from a common origin; these proteins are ''Homology (biology), homologous''. Phylogenetics, Phylogenetic analysis of proteins has revealed how proteins evolve and change their structure and function over time.
Evolutionary rate. Using the amino acid sequences of hemoglobin and cytochrome c from multiple species, scientists were able to derive estimations of protein evolution rates. Each protein has its own rate, and that rate is relatively constant across phylogenies (i.e., hemoglobin does not evolve at the same rate as cytochrome c, but hemoglobins from humans, mice, etc. do have comparable rates of evolution). Not all regions within a protein mutate at the same rate; functionally important areas mutate more slowly and amino acid substitutions involving similar amino acids occurs more often than dissimilar substitutions. Overall, the level of polymorphisms in proteins seems to be fairly constant. Several species (including humans, fruit flies, and mice) have similar levels of protein polymorphism.
Functional evolution. Numerous enzymes and other proteins have been shown to change their function over the course of evolution. For example, ribonucleotide reductase (RNR) is known from thousands of organisms and has evolved a multitude of structural and functional variants. Class I RNRs use a ferritin subunit and differ by the metal they use as cofactors. In class II RNRs, the thiyl radical is generated using an adenosylcobalamin cofactor and these enzymes do not require additional subunits (as opposed to class I which do). In class III RNRs, the thiyl radical is generated using S-Adenosyl methionine, S-adenosylmethionine bound to a [
molecule
A molecule is a group of two or more atoms held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions which satisfy this criterion. In quantum physics, organic chemistry, and bioche ...
s such as DNA, RNA, and protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
s across generations. The field of molecular evolution uses principles of evolutionary biology
Evolutionary biology is the subfield of biology that studies the evolutionary processes ( natural selection, common descent, speciation) that produced the diversity of life on Earth. It is also defined as the study of the history of life ...
and population genetics
Population genetics is a subfield of genetics that deals with genetic differences within and between populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as Adaptation (biology), adaptation, ...
to explain patterns in these changes. Major topics in molecular evolution concern the rates and impacts of single nucleotide changes, neutral evolution vs. natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the heritable traits characteristic of a population over generations. Cha ...
, origins of new genes, the genetic nature of complex traits, the genetic basis of speciation, evolution of development, and ways that evolutionary forces influence genomic and phenotypic changes.
History
The history of molecular evolution starts in the early 20th century with comparativebiochemistry
Biochemistry or biological chemistry is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology and ...
, and the use of "fingerprinting" methods such as immune assays, gel electrophoresis and paper chromatography
Paper chromatography is an analytical method used to separate coloured chemicals or substances. It is now primarily used as a teaching tool, having been replaced in the laboratory by other chromatography methods such as thin-layer chromatography ...
in the 1950s to explore homologous proteins.
The field of molecular evolution came into its own in the 1960s and 1970s, following the rise of molecular biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physi ...
. The advent of protein sequencing
Protein sequencing is the practical process of determining the amino acid sequence of all or part of a protein or peptide. This may serve to identify the protein or characterize its post-translational modifications. Typically, partial sequencing o ...
allowed molecular biologists to create phylogenies based on sequence comparison, and to use the differences between homologous sequences as a molecular clock to estimate the time since the last universal common ancestor
The last universal common ancestor (LUCA) is the most recent population from which all organisms now living on Earth share common descent—the most recent common ancestor of all current life on Earth. This includes all cellular organisms; th ...
. In the late 1960s, the neutral theory of molecular evolution
The neutral theory of molecular evolution holds that most evolutionary changes occur at the molecular level, and most of the variation within and between species are due to random genetic drift of mutant alleles that are selectively neutral. The ...
provided a theoretical basis for the molecular clock, though both the clock and the neutral theory were controversial, since most evolutionary biologists held strongly to panselectionism, with natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the heritable traits characteristic of a population over generations. Cha ...
as the only important cause of evolutionary change. After the 1970s, nucleic acid sequencing allowed molecular evolution to reach beyond proteins to highly conserved ribosomal RNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosom ...
sequences, the foundation of a reconceptualization of the early history of life
The history of life on Earth traces the processes by which living and fossil organisms evolved, from the earliest emergence of life to present day. Earth formed about 4.5 billion years ago (abbreviated as ''Ga'', for ''gigaannum'') and evide ...
.
Forces in molecular evolution
The content and structure of a genome is the product of the molecular and population genetic forces which act upon that genome. Novel genetic variants will arise throughmutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA replication, DNA or viral repl ...
and will spread and be maintained in populations due to genetic drift or natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the heritable traits characteristic of a population over generations. Cha ...
.
Mutation
 Mutations are permanent, transmissible changes to the
Mutations are permanent, transmissible changes to the genetic material
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main clas ...
( DNA or RNA) of a cell
Cell most often refers to:
* Cell (biology), the functional basic unit of life
Cell may also refer to:
Locations
* Monastic cell, a small room, hut, or cave in which a religious recluse lives, alternatively the small precursor of a monastery ...
or virus
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea.
Since Dmitri Ivanovsk ...
. Mutations result from errors in DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritanc ...
during cell division
Cell division is the process by which a parent cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukaryotes, there ar ...
and by exposure to radiation, chemicals, and other environmental stressors, or viruses
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea.
Since Dmitri Ivanovsky's ...
and transposable elements
A transposable element (TE, transposon, or jumping gene) is a nucleic acid sequence in DNA that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size. Trans ...
. Most mutations that occur are single nucleotide polymorphisms which modify single bases of the DNA sequence, resulting in point mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequence ...
s. Other types of mutations modify larger segments of DNA and can cause duplications, insertions, deletions, inversions, and translocations.
Most organisms display a strong bias in the types of mutations that occur with strong influence in GC-content. Transitions (A ↔ G or C ↔ T) are more common than transversion
Transversion, in molecular biology, refers to a point mutation in DNA in which a single (two ring) purine ( A or G) is changed for a (one ring) pyrimidine ( T or C), or vice versa. A transversion can be spontaneous, or it can be caused by i ...
s ( purine (adenine or guanine)) ↔ pyrimidine (cytosine or thymine, or in RNA, uracil)) and are less likely to alter amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha a ...
sequences of protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
s.
Mutations are stochastic and typically occur randomly across genes. Mutation rates for single nucleotide sites for most organisms are very low, roughly 10−9 to 10−8 per site per generation, though some viruses have higher mutation rates on the order of 10−6 per site per generation. Among these mutations, some will be neutral or beneficial and will remain in the genome unless lost via genetic drift, and others will be detrimental and will be eliminated from the genome by natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the heritable traits characteristic of a population over generations. Cha ...
.
Because mutations are extremely rare, they accumulate very slowly across generations. While the number of mutations which appears in any single generation may vary, over very long time periods they will appear to accumulate at a regular pace. Using the mutation rate per generation and the number of nucleotide differences between two sequences, divergence times can be estimated effectively via the molecular clock.
Recombination
Gene conversion
Gene conversion is a type of recombination that is the product ofDNA repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA da ...
where nucleotide damage is corrected using an homologous genomic region as a template. Damaged bases are first excised, the damaged strand is then aligned with an undamaged homolog, and DNA synthesis repairs the excised region using the undamaged strand as a guide. Gene conversion is often responsible for homogenizing sequences of duplicate genes over long time periods, reducing nucleotide divergence.
Genetic drift
Genetic drift is the change of allele frequencies from one generation to the next due to stochastic effects ofrandom sampling
In statistics, quality assurance, and survey methodology, sampling is the selection of a subset (a statistical sample) of individuals from within a statistical population to estimate characteristics of the whole population. Statisticians attemp ...
in finite populations. Some existing variants have no effect on fitness and may increase or decrease in frequency simply due to chance. "Nearly neutral" variants whose selection coefficient
In population genetics, a selection coefficient, usually denoted by the letter ''s'', is a measure of differences in relative fitness. Selection coefficients are central to the quantitative description of evolution, since fitness differences deter ...
is close to a threshold value of 1 / the effective population size
The effective population size (''N'e'') is a number that, in some simplified scenarios, corresponds to the number of breeding individuals in the population. More generally, ''N'e'' is the number of individuals that an idealised population w ...
will also be affected by chance as well as by selection and mutation. Many genomic features have been ascribed to accumulation of nearly neutral detrimental mutations as a result of small effective population sizes. With a smaller effective population size, a larger variety of mutations will behave as if they are neutral due to inefficiency of selection.
Selection
Selection occurs when organisms with greater fitness, i.e. greater ability to survive or reproduce, are favored in subsequent generations, thereby increasing the instance of underlying genetic variants in a population. Selection can be the product of natural selection, artificial selection, or sexual selection.Natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the heritable traits characteristic of a population over generations. Cha ...
is any selective process that occurs due to the fitness of an organism to its environment. In contrast sexual selection
Sexual selection is a mode of natural selection in which members of one biological sex choose mates of the other sex to mate with (intersexual selection), and compete with members of the same sex for access to members of the opposite sex ( ...
is a product of mate choice and can favor the spread of genetic variants which act counter to natural selection but increase desirability to the opposite sex or increase mating success. Artificial selection, also known as selective breeding, is imposed by an outside entity, typically humans, in order to increase the frequency of desired traits.
The principles of population genetics apply similarly to all types of selection, though in fact each may produce distinct effects due to clustering of genes with different functions in different parts of the genome, or due to different properties of genes in particular functional classes. For instance, sexual selection could be more likely to affect molecular evolution of the sex chromosomes due to clustering of sex specific genes on the X, Y, Z or W.
Intragenomic conflict
Selection can operate at the gene level at the expense of organismal fitness, resulting inintragenomic conflict
Intragenomic conflict refers to the evolutionary phenomenon where genes have phenotypic effects that promote their own transmission in detriment of the transmission of other genes that reside in the same genome. The selfish gene theory postulates ...
. This is because there can be a selective advantage for selfish genetic elements in spite of a host cost. Examples of such selfish elements include transposable elements, meiotic drivers, killer X chromosomes, selfish mitochondria, and self-propagating introns.
Genome architecture
Genome size
Genome size is influenced by the amount of repetitive DNA as well as number of genes in an organism. TheC-value paradox
C-value is the amount, in picograms, of DNA contained within a haploid nucleus (e.g. a gamete) or one half the amount in a diploid somatic cell of a eukaryotic organism. In some cases (notably among diploid organisms), the terms C-value and gen ...
refers to the lack of correlation between organism 'complexity' and genome size. Explanations for the so-called paradox are two-fold. First, repetitive genetic elements can comprise large portions of the genome for many organisms, thereby inflating DNA content of the haploid genome. Secondly, the number of genes is not necessarily indicative of the number of developmental stages or tissue types in an organism. An organism with few developmental stages or tissue types may have large numbers of genes that influence non-developmental phenotypes, inflating gene content relative to developmental gene families.
Neutral explanations for genome size suggest that when population sizes are small, many mutations become nearly neutral. Hence, in small populations repetitive content and other 'junk' DNA can accumulate without placing the organism at a competitive disadvantage. There is little evidence to suggest that genome size is under strong widespread selection in multicellular eukaryotes. Genome size, independent of gene content, correlates poorly with most physiological traits and many eukaryotes, including mammals, harbor very large amounts of repetitive DNA.
However, birds likely have experienced strong selection for reduced genome size, in response to changing energetic needs for flight. Birds, unlike humans, produce nucleated red blood cells, and larger nuclei lead to lower levels of oxygen transport. Bird metabolism is far higher than that of mammals, due largely to flight, and oxygen needs are high. Hence, most birds have small, compact genomes with few repetitive elements. Indirect evidence suggests that non-avian theropod dinosaur ancestors of modern birds also had reduced genome sizes, consistent with endothermy and high energetic needs for running speed. Many bacteria have also experienced selection for small genome size, as time of replication and energy consumption are so tightly correlated with fitness.
Repetitive elements
Transposable elements
A transposable element (TE, transposon, or jumping gene) is a nucleic acid sequence in DNA that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size. Trans ...
are self-replicating, selfish genetic elements which are capable of proliferating within host genomes. Many transposable elements are related to viruses, and share several proteins in common....
Chromosome number and organization
The number of chromosomes in an organism's genome also does not necessarily correlate with the amount of DNA in its genome. The ant ''Myrmecia pilosula'' has only a single pair of chromosomes whereas the Adders-tongue fern ''Ophioglossum
''Ophioglossum'', the adder's-tongue ferns, is a genus of about 50 species of ferns in the family Ophioglossaceae. The name ''Ophioglossum'' comes from the Greek meaning "snake-tongue".
reticulatum'' has up to 1260 chromosomes. Cilliate genomes house each gene in individual chromosomes, resulting in a genome which is not physically linked. Reduced linkage through creation of additional chromosomes should effectively increase the efficiency of selection.
Changes in chromosome number can play a key role in speciation, as differing chromosome numbers can serve as a barrier to reproduction in hybrids. Human chromosome 2
Chromosome 2 is one of the twenty-three pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 2 is the second-largest human chromosome, spanning more than 242 million base pairs and representing almost e ...
was created from a fusion of two chimpanzee chromosomes and still contains central telomeres
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes. Although there are different architectures, telomeres, in a broad sense, are a widespread genetic feature mos ...
as well as a vestigial second centromere
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers ...
. Polyploidy, especially allopolyploidy, which occurs often in plants, can also result in reproductive incompatibilities with parental species. ''Agrodiatus'' blue butterflies have diverse chromosome numbers ranging from n=10 to n=134 and additionally have one of the highest rates of speciation identified to date.
Gene content and distribution
Different organisms house different numbers of genes within their genomes as well as different patterns in the distribution of genes throughout the genome. Some organisms, such as most bacteria, ''Drosophila'', and ''Arabidopsis'' have particularly compact genomes with little repetitive content or non-coding DNA. Other organisms, like mammals or maize, have large amounts of repetitive DNA, long introns, and substantial spacing between different genes. The content and distribution of genes within the genome can influence the rate at which certain types of mutations occur and can influence the subsequent evolution of different species. Genes with longerintrons
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene ...
are more likely to recombine due to increased physical distance over the coding sequence. As such, long introns may facilitate ectopic recombination
Ectopic recombination is an atypical form of recombination in which crossing over occurs at non-homologous, rather than along homologous, loci. //This needs to be edited, as it is 1)incorrect and 2)contradicts what's written below, namely the ne ...
, and result in higher rates of new gene formation.
Organelles
In addition to the nuclear genome, endosymbiont organelles contain their own genetic material typically as circular plasmids. Mitochondrial and chloroplast DNA varies across taxa, but membrane-bound proteins, especially electron transport chain constituents are most often encoded in the organelle. Chloroplasts and mitochondria are maternally inherited in most species, as the organelles must pass through the egg. In a rare departure, some species of mussels are known to inherit mitochondria from father to son.Origins of new genes
Newgene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
s arise from several different genetic mechanisms including gene duplication, de novo origination, retrotransposition, chimeric gene formation, recruitment of non-coding sequence, and gene truncation.
Gene duplication initially leads to redundancy. However, duplicated gene sequences can mutate to develop new functions or specialize so that the new gene performs a subset of the original ancestral functions. In addition to duplicating whole genes, sometimes only a domain or part of a protein is duplicated so that the resulting gene is an elongated version of the parental gene.
Retrotransposition creates new genes by copying mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
to DNA and inserting it into the genome. Retrogenes often insert into new genomic locations, and often develop new expression patterns and functions.
Chimeric genes form when duplication, deletion, or incomplete retrotransposition combine portions of two different coding sequences to produce a novel gene sequence. Chimeras often cause regulatory changes and can shuffle protein domains to produce novel adaptive functions.
''De novo'' gene birth can also give rise to new genes from previously non-coding DNA
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules (e.g. transfer RNA, microRNA, piRNA, ribosomal RNA, and regula ...
. For instance, Levine and colleagues reported the origin of five new genes in the ''D. melanogaster'' genome from noncoding DNA. Similar de novo origin of genes has been also shown in other organisms such as yeast, rice and humans. De novo genes may evolve from transcripts that are already expressed at low levels. Mutation of a stop codon
In molecular biology (specifically protein biosynthesis), a stop codon (or termination codon) is a codon (nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in mess ...
to a regular codon or a frameshift may cause an extended protein that includes a previously non-coding sequence. The formation of novel genes from scratch typically can not occur within genomic regions of high gene density. The essential events for de novo formation of genes is recombination/mutation which includes insertions, deletions, and inversions. These events are tolerated if the consequence of these genetic events does not interfere in cellular activities. Most genomes comprise prophages wherein genetic modifications do not, in general, affect the host genome propagation. Hence, there is higher probability of genetic modifications, in regions such as prophages, which is proportional to the probability of de novo formation of genes.
''De novo'' evolution of genes can also be simulated in the laboratory. For example, semi-random gene sequences can be selected for specific functions. More specifically, they selected sequences from a library that could complement a gene deletion in '' E. coli''. The deleted gene encodes ferric enterobactin esterase (Fes), which releases iron from an iron chelator, enterobactin
Enterobactin (also known as enterochelin) is a high affinity siderophore that acquires iron for microbial systems. It is primarily found in Gram-negative bacteria, such as ''Escherichia coli'' and '' Salmonella typhimurium''.
Enterobactin is t ...
. While Fes is a 400 amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha a ...
protein, the newly selected gene was only 100 amino acids in length and unrelated in sequence to Fes.
''In vitro'' molecular evolution experiments
Principles of molecular evolution can be discovered and tested using laboratory experimentation. This usually involves the cloning and ''in vitro'' modification of genes and proteins outside cells. Since the pioneering work of Sol Spiegelmann in 1967 ef involving RNA that replicates itself with the aid of an enzyme extracted from the Qß virus ef several groups (such as Kramers efand Biebricher/Luce/Eigen ef studied mini and micro variants of this RNA in the 1970s and 1980s that replicate on the timescale of seconds to a minute, allowing hundreds of generations with large population sizes (e.g. 10^14 sequences) to be followed in a single day of experimentation. The chemical kinetic elucidation of the detailed mechanism of replication ef, refmeant that this type of system was the first molecular evolution system that could be fully characterised on the basis of physical chemical kinetics, later allowing the first models of the genotype to phenotype map based on sequence dependent RNA folding and refolding to be produced ef, ref Subject to maintaining the function of the multicomponent Qß enzyme, chemical conditions could be varied significantly, in order to study the influence of changing environments and selection pressures ef Experiments with ''in vitro'' RNA quasi species included the characterisation of the error threshold for information in molecular evolution ef the discovery of ''de novo'' evolution efleading to diverse replicating RNA species and the discovery of spatial travelling waves as ideal molecular evolution reactors ef, ref Later experiments employed novel combinations of enzymes to elucidate novel aspects of interacting molecular evolution involving population dependent fitness, including work with artificially designed molecular predator prey and cooperative systems of multiple RNA and DNA ef, ref Special evolution reactors were designed for these studies, starting with serial transfer machines, flow reactors such as cell-stat machines, capillary reactors, and microreactors including line flow reactors and gel slice reactors. These studies were accompanied by theoretical developments and simulations involving RNA folding and replication kinetics that elucidated the importance of the correlation structure between distance in sequence space and fitness changes ef including the role of neutral networks and structural ensembles in evolutionary optimisation.in vitro protein function evolution
Mutagenic hot spots in enzymes can be identified usingNMR spectroscopy
Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy or magnetic resonance spectroscopy (MRS), is a spectroscopic technique to observe local magnetic fields around atomic nuclei. The sample is placed in a magnetic fie ...
. In a proof-of-concept study, Bhattacharya and colleagues converted myoglobin, a non-enzymatic oxygen storage protein, into a highly efficient Kemp eliminase using only three mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA replication, DNA or viral repl ...
s. This demonstrates that only few mutations are needed to radically change the function of a protein.
Molecular phylogenetics
Molecular systematics is the product of the traditional fields of systematics andmolecular genetics
Molecular genetics is a sub-field of biology that addresses how differences in the structures or expression of DNA molecules manifests as variation among organisms. Molecular genetics often applies an "investigative approach" to determine the ...
. It uses DNA, RNA, or protein sequences to resolve questions in systematics, i.e. about their correct scientific classification
Taxonomy is the practice and science of categorization or classification.
A taxonomy (or taxonomical classification) is a scheme of classification, especially a hierarchical classification, in which things are organized into groups or types. ...
or taxonomy
Taxonomy is the practice and science of categorization or classification.
A taxonomy (or taxonomical classification) is a scheme of classification, especially a hierarchical classification, in which things are organized into groups or types. ...
from the point of view of evolutionary biology
Evolutionary biology is the subfield of biology that studies the evolutionary processes ( natural selection, common descent, speciation) that produced the diversity of life on Earth. It is also defined as the study of the history of life ...
.
Molecular systematics has been made possible by the availability of techniques for DNA sequencing, which allow the determination of the exact sequence of nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecule ...
s or ''bases'' in either DNA or RNA. At present it is still a long and expensive process to sequence the entire genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding g ...
of an organism, and this has been done for only a few species. However, it is quite feasible to determine the sequence of a defined area of a particular chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins are ...
. Typical molecular systematic analyses require the sequencing of around 1000 base pairs.
The driving forces of evolution
Depending on the relative importance assigned to the various forces of evolution, three perspectives provide evolutionary explanations for molecular evolution. Selectionist hypotheses argue that selection is the driving force of molecular evolution. While acknowledging that many mutations are neutral, selectionists attribute changes in the frequencies of neutral alleles tolinkage disequilibrium
In population genetics, linkage disequilibrium (LD) is the non-random association of alleles at different loci in a given population. Loci are said to be in linkage disequilibrium when the frequency of association of their different alleles is h ...
with other loci that are under selection, rather than to random genetic drift. Biases in codon usage are usually explained with reference to the ability of even weak selection to shape molecular evolution.
Neutralist hypotheses emphasize the importance of mutation, purifying selection, and random genetic drift. The introduction of the neutral theory by Kimura, quickly followed by King
King is the title given to a male monarch in a variety of contexts. The female equivalent is queen, which title is also given to the consort of a king.
*In the context of prehistory, antiquity and contemporary indigenous peoples, the tit ...
and Jukes Jukes is a surname. Notable people with the surname include:
* Andrew Jukes (theologian) (1815–1901)
*Andrew Jukes (missionary) (1847–1931), Anglican missionary
* Betty Jukes (1910–2006), British sculptor
* Bill Jukes (c.1883–1939), English ...
' own findings, led to a fierce debate about the relevance of neodarwinism at the molecular level. The Neutral theory of molecular evolution
The neutral theory of molecular evolution holds that most evolutionary changes occur at the molecular level, and most of the variation within and between species are due to random genetic drift of mutant alleles that are selectively neutral. The ...
proposes that most mutations in DNA are at locations not important to function or fitness. These neutral changes drift towards fixation within a population. Positive changes will be very rare, and so will not greatly contribute to DNA polymorphisms. Deleterious mutations do not contribute much to DNA diversity because they negatively affect fitness and so are removed from the gene pool before long. This theory provides a framework for the molecular clock. The fate of neutral mutations are governed by genetic drift, and contribute to both nucleotide polymorphism and fixed differences between species.The nearly neutral theory expanded the neutralist perspective, suggesting that several mutations are nearly neutral, which means both random drift and natural selection is relevant to their dynamics.
In the strictest sense, the neutral theory is not accurate. Subtle changes in DNA very often have effects, but sometimes these effects are too small for natural selection to act on. Even synonymous mutations are not necessarily neutral because there is not a uniform amount of each codon. The nearly neutral theory expanded the neutralist perspective, suggesting that several mutations are nearly neutral, which means both random drift and natural selection is relevant to their dynamics. The main difference between the neutral theory and nearly neutral theory is that the latter focuses on weak selection, not strictly neutral.
Another concept is constructive neutral evolution Constructive neutral evolution (CNE) is a theory that seeks to explain how complex systems can evolve through neutral transitions and spread through a population by chance fixation ( genetic drift). Constructive neutral evolution is a competitor fo ...
(CNE), which explains that complex systems can emerge and spread into a population through neutral transitions with the principles of excess capacity, presuppression, and ratcheting, and it has been applied in areas ranging from the origins of the spliceosome to the complex interdependence of Microbial consortium, microbial communities.
Mutationists hypotheses emphasize random drift and biases in mutation patterns. Sueoka was the first to propose a modern mutationist view. He proposed that the variation in GC content was not the result of positive selection, but a consequence of the GC mutational pressure.
Protein evolution
Whilegenome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding g ...
s store information and accumulate mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA replication, DNA or viral repl ...
s, proteins are the active products of gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
s. Hence the evolution of protein function is critical to understand molecular evolution.  Evolution of proteins is studied by Sequence alignment, comparing the sequences and Protein structure, structures of proteins from many organisms. Similar sequences/structures indicating that the proteins diverged from a common origin; these proteins are ''Homology (biology), homologous''. Phylogenetics, Phylogenetic analysis of proteins has revealed how proteins evolve and change their structure and function over time.
Evolutionary rate. Using the amino acid sequences of hemoglobin and cytochrome c from multiple species, scientists were able to derive estimations of protein evolution rates. Each protein has its own rate, and that rate is relatively constant across phylogenies (i.e., hemoglobin does not evolve at the same rate as cytochrome c, but hemoglobins from humans, mice, etc. do have comparable rates of evolution). Not all regions within a protein mutate at the same rate; functionally important areas mutate more slowly and amino acid substitutions involving similar amino acids occurs more often than dissimilar substitutions. Overall, the level of polymorphisms in proteins seems to be fairly constant. Several species (including humans, fruit flies, and mice) have similar levels of protein polymorphism.
Functional evolution. Numerous enzymes and other proteins have been shown to change their function over the course of evolution. For example, ribonucleotide reductase (RNR) is known from thousands of organisms and has evolved a multitude of structural and functional variants. Class I RNRs use a ferritin subunit and differ by the metal they use as cofactors. In class II RNRs, the thiyl radical is generated using an adenosylcobalamin cofactor and these enzymes do not require additional subunits (as opposed to class I which do). In class III RNRs, the thiyl radical is generated using S-Adenosyl methionine, S-adenosylmethionine bound to a [
Evolution of proteins is studied by Sequence alignment, comparing the sequences and Protein structure, structures of proteins from many organisms. Similar sequences/structures indicating that the proteins diverged from a common origin; these proteins are ''Homology (biology), homologous''. Phylogenetics, Phylogenetic analysis of proteins has revealed how proteins evolve and change their structure and function over time.
Evolutionary rate. Using the amino acid sequences of hemoglobin and cytochrome c from multiple species, scientists were able to derive estimations of protein evolution rates. Each protein has its own rate, and that rate is relatively constant across phylogenies (i.e., hemoglobin does not evolve at the same rate as cytochrome c, but hemoglobins from humans, mice, etc. do have comparable rates of evolution). Not all regions within a protein mutate at the same rate; functionally important areas mutate more slowly and amino acid substitutions involving similar amino acids occurs more often than dissimilar substitutions. Overall, the level of polymorphisms in proteins seems to be fairly constant. Several species (including humans, fruit flies, and mice) have similar levels of protein polymorphism.
Functional evolution. Numerous enzymes and other proteins have been shown to change their function over the course of evolution. For example, ribonucleotide reductase (RNR) is known from thousands of organisms and has evolved a multitude of structural and functional variants. Class I RNRs use a ferritin subunit and differ by the metal they use as cofactors. In class II RNRs, the thiyl radical is generated using an adenosylcobalamin cofactor and these enzymes do not require additional subunits (as opposed to class I which do). In class III RNRs, the thiyl radical is generated using S-Adenosyl methionine, S-adenosylmethionine bound to a [Relation to nucleic acid evolution
Protein evolution is inescapably tied to changes and selection of DNA polymorphisms and mutations because protein sequences change in response to alterations in the DNA sequence. Amino acid sequences and nucleic acid sequences do not mutate at the same rate. Due to the degenerate nature of DNA, bases can change without affecting the amino acid sequence. For example, there are six codons that code for leucine. Thus, despite the difference in mutation rates, it is essential to incorporate nucleic acid evolution into the discussion of protein evolution. At the end of the 1960s, two groups of scientists—Kimura (1968) and King and Jukes (1969)—independently proposed that a majority of the evolutionary changes observed in proteins were neutral. Since then, the neutral theory has been expanded upon and debated.Discordance with morphological evolution
There are sometimes discordances between molecular and Morphology (biology), morphological evolution, which are reflected in molecular and morphological systematic studies, especially of bacteria, archaea and eukaryotic microbes. These discordances can be categorized as two types: (i) one morphology, multiple lineages (e.g. convergent evolution, morphological convergence, cryptic species) and (ii) one lineage, multiple morphologies (e.g. phenotypic plasticity, multiple biological life cycle, life-cycle stages). Neutral evolution possibly could explain the incongruences in some cases.Journals and societies
The Society for Molecular Biology and Evolution publishes the journals "Molecular Biology and Evolution" and "Genome Biology and Evolution" and holds an annual international meeting. Other journals dedicated to molecular evolution include ''Journal of Molecular Evolution'' and ''Molecular Phylogenetics and Evolution''. Research in molecular evolution is also published in journals of genetics,molecular biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physi ...
, genomics, systematics, and evolutionary biology
Evolutionary biology is the subfield of biology that studies the evolutionary processes ( natural selection, common descent, speciation) that produced the diversity of life on Earth. It is also defined as the study of the history of life ...
.
See also
* Abiogenesis * Adaptor proteins, vesicular transport#Evolutionary considerations, Adaptor protein evolution * Comparative phylogenetics * Evolution * E. coli long-term evolution experiment, ''E. coli'' long-term evolution experiment * Evolutionary physiology * Evolution of dietary antioxidants * Genomic organization * Genetic drift * Genome evolution * Heterotachy * History of molecular evolution * Horizontal gene transfer * Human evolution * Molecular clock * Molecular paleontology *Neutral theory of molecular evolution
The neutral theory of molecular evolution holds that most evolutionary changes occur at the molecular level, and most of the variation within and between species are due to random genetic drift of mutant alleles that are selectively neutral. The ...
* Nucleotide diversity
* Maximum parsimony (phylogenetics), Parsimony
* Population genetics
* Selection (biology), Selection
References
Further reading
* * * * * {{Authority control Molecular evolution,