Transcription (biology) on:
[Wikipedia]
[Google]
[Amazon]
Transcription is the process of copying a segment of
 Setting up for transcription in mammals is regulated by many cis-regulatory elements, including core promoter and promoter-proximal elements that are located near the transcription start sites of genes. Core promoters combined with general transcription factors are sufficient to direct transcription initiation, but generally have low basal activity. Other important cis-regulatory modules are localized in DNA regions that are distant from the transcription start sites. These include enhancers, silencers, insulators and tethering elements. Among this constellation of elements, enhancers and their associated transcription factors have a leading role in the initiation of gene transcription. An enhancer localized in a DNA region distant from the promoter of a gene can have a very large effect on gene transcription, with some genes undergoing up to 100-fold increased transcription due to an activated enhancer.
Enhancers are regions of the genome that are major gene-regulatory elements. Enhancers control cell-type-specific gene transcription programs, most often by looping through long distances to come in physical proximity with the promoters of their target genes. While there are hundreds of thousands of enhancer DNA regions, for a particular type of tissue only specific enhancers are brought into proximity with the promoters that they regulate. In a study of brain cortical neurons, 24,937 loops were found, bringing enhancers to their target promoters. Multiple enhancers, each often at tens or hundred of thousands of nucleotides distant from their target genes, loop to their target gene promoters and can coordinate with each other to control transcription of their common target gene.
The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene. The loop is stabilized by a dimer of a connector protein (e.g. dimer of CTCF or YY1), with one member of the dimer anchored to its binding motif on the enhancer and the other member anchored to its binding motif on the promoter (represented by the red zigzags in the illustration). Several cell function specific transcription factors (there are about 1,600 transcription factors in a human cell) generally bind to specific motifs on an enhancer and a small combination of these enhancer-bound transcription factors, when brought close to a promoter by a DNA loop, govern level of transcription of the target gene. Mediator (a complex usually consisting of about 26 proteins in an interacting structure) communicates regulatory signals from enhancer DNA-bound transcription factors directly to the RNA polymerase II (pol II) enzyme bound to the promoter.
Enhancers, when active, are generally transcribed from both strands of DNA with RNA polymerases acting in two different directions, producing two enhancer RNAs (eRNAs) as illustrated in the Figure. An inactive enhancer may be bound by an inactive transcription factor. Phosphorylation of the transcription factor may activate it and that activated transcription factor may then activate the enhancer to which it is bound (see small red star representing phosphorylation of transcription factor bound to enhancer in the illustration). An activated enhancer begins transcription of its RNA before activating transcription of messenger RNA from its target gene.
Setting up for transcription in mammals is regulated by many cis-regulatory elements, including core promoter and promoter-proximal elements that are located near the transcription start sites of genes. Core promoters combined with general transcription factors are sufficient to direct transcription initiation, but generally have low basal activity. Other important cis-regulatory modules are localized in DNA regions that are distant from the transcription start sites. These include enhancers, silencers, insulators and tethering elements. Among this constellation of elements, enhancers and their associated transcription factors have a leading role in the initiation of gene transcription. An enhancer localized in a DNA region distant from the promoter of a gene can have a very large effect on gene transcription, with some genes undergoing up to 100-fold increased transcription due to an activated enhancer.
Enhancers are regions of the genome that are major gene-regulatory elements. Enhancers control cell-type-specific gene transcription programs, most often by looping through long distances to come in physical proximity with the promoters of their target genes. While there are hundreds of thousands of enhancer DNA regions, for a particular type of tissue only specific enhancers are brought into proximity with the promoters that they regulate. In a study of brain cortical neurons, 24,937 loops were found, bringing enhancers to their target promoters. Multiple enhancers, each often at tens or hundred of thousands of nucleotides distant from their target genes, loop to their target gene promoters and can coordinate with each other to control transcription of their common target gene.
The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene. The loop is stabilized by a dimer of a connector protein (e.g. dimer of CTCF or YY1), with one member of the dimer anchored to its binding motif on the enhancer and the other member anchored to its binding motif on the promoter (represented by the red zigzags in the illustration). Several cell function specific transcription factors (there are about 1,600 transcription factors in a human cell) generally bind to specific motifs on an enhancer and a small combination of these enhancer-bound transcription factors, when brought close to a promoter by a DNA loop, govern level of transcription of the target gene. Mediator (a complex usually consisting of about 26 proteins in an interacting structure) communicates regulatory signals from enhancer DNA-bound transcription factors directly to the RNA polymerase II (pol II) enzyme bound to the promoter.
Enhancers, when active, are generally transcribed from both strands of DNA with RNA polymerases acting in two different directions, producing two enhancer RNAs (eRNAs) as illustrated in the Figure. An inactive enhancer may be bound by an inactive transcription factor. Phosphorylation of the transcription factor may activate it and that activated transcription factor may then activate the enhancer to which it is bound (see small red star representing phosphorylation of transcription factor bound to enhancer in the illustration). An activated enhancer begins transcription of its RNA before activating transcription of messenger RNA from its target gene.
 Transcription regulation at about 60% of promoters is also controlled by methylation of cytosines within CpG dinucleotides (where 5' cytosine is followed by 3' guanine or CpG sites). 5-methylcytosine (5-mC) is a methylated form of the
Transcription regulation at about 60% of promoters is also controlled by methylation of cytosines within CpG dinucleotides (where 5' cytosine is followed by 3' guanine or CpG sites). 5-methylcytosine (5-mC) is a methylated form of the  As noted in the previous section, transcription factors are proteins that bind to specific DNA sequences in order to regulate the expression of a gene. The binding sequence for a transcription factor in DNA is usually about 10 or 11 nucleotides long. As summarized in 2009, Vaquerizas et al. indicated there are approximately 1,400 different transcription factors encoded in the human genome by genes that constitute about 6% of all human protein encoding genes. About 94% of transcription factor binding sites (TFBSs) that are associated with signal-responsive genes occur in enhancers while only about 6% of such TFBSs occur in promoters.
EGR1 protein is a particular transcription factor that is important for regulation of methylation of CpG islands. An EGR1 transcription factor binding site is frequently located in enhancer or promoter sequences. There are about 12,000 binding sites for EGR1 in the mammalian genome and about half of EGR1 binding sites are located in promoters and half in enhancers. The binding of EGR1 to its target DNA binding site is insensitive to cytosine methylation in the DNA.
While only small amounts of EGR1 transcription factor protein are detectable in cells that are un-stimulated, translation of the ''EGR1'' gene into protein at one hour after stimulation is drastically elevated. Production of EGR1 transcription factor proteins, in various types of cells, can be stimulated by growth factors, neurotransmitters, hormones, stress and injury. In the brain, when neurons are activated, EGR1 proteins are up-regulated and they bind to (recruit) the pre-existing TET1 enzymes that are produced in high amounts in neurons. TET enzymes can catalyse demethylation of 5-methylcytosine. When EGR1 transcription factors bring TET1 enzymes to EGR1 binding sites in promoters, the TET enzymes can demethylate the methylated CpG islands at those promoters. Upon demethylation, these promoters can then initiate transcription of their target genes. Hundreds of genes in neurons are differentially expressed after neuron activation through EGR1 recruitment of TET1 to methylated regulatory sequences in their promoters.
The methylation of promoters is also altered in response to signals. The three mammalian DNA methyltransferasess (DNMT1, DNMT3A, and DNMT3B) catalyze the addition of methyl groups to cytosines in DNA. While DNMT1 is a maintenance methyltransferase, DNMT3A and DNMT3B can carry out new methylations. There are also two splice protein isoforms produced from the ''DNMT3A'' gene: DNA methyltransferase proteins DNMT3A1 and DNMT3A2.
The splice isoform DNMT3A2 behaves like the product of a classical immediate-early gene and, for instance, it is robustly and transiently produced after neuronal activation. Where the DNA methyltransferase isoform DNMT3A2 binds and adds methyl groups to cytosines appears to be determined by histone post translational modifications.
On the other hand, neural activation causes degradation of DNMT3A1 accompanied by reduced methylation of at least one evaluated targeted promoter.
As noted in the previous section, transcription factors are proteins that bind to specific DNA sequences in order to regulate the expression of a gene. The binding sequence for a transcription factor in DNA is usually about 10 or 11 nucleotides long. As summarized in 2009, Vaquerizas et al. indicated there are approximately 1,400 different transcription factors encoded in the human genome by genes that constitute about 6% of all human protein encoding genes. About 94% of transcription factor binding sites (TFBSs) that are associated with signal-responsive genes occur in enhancers while only about 6% of such TFBSs occur in promoters.
EGR1 protein is a particular transcription factor that is important for regulation of methylation of CpG islands. An EGR1 transcription factor binding site is frequently located in enhancer or promoter sequences. There are about 12,000 binding sites for EGR1 in the mammalian genome and about half of EGR1 binding sites are located in promoters and half in enhancers. The binding of EGR1 to its target DNA binding site is insensitive to cytosine methylation in the DNA.
While only small amounts of EGR1 transcription factor protein are detectable in cells that are un-stimulated, translation of the ''EGR1'' gene into protein at one hour after stimulation is drastically elevated. Production of EGR1 transcription factor proteins, in various types of cells, can be stimulated by growth factors, neurotransmitters, hormones, stress and injury. In the brain, when neurons are activated, EGR1 proteins are up-regulated and they bind to (recruit) the pre-existing TET1 enzymes that are produced in high amounts in neurons. TET enzymes can catalyse demethylation of 5-methylcytosine. When EGR1 transcription factors bring TET1 enzymes to EGR1 binding sites in promoters, the TET enzymes can demethylate the methylated CpG islands at those promoters. Upon demethylation, these promoters can then initiate transcription of their target genes. Hundreds of genes in neurons are differentially expressed after neuron activation through EGR1 recruitment of TET1 to methylated regulatory sequences in their promoters.
The methylation of promoters is also altered in response to signals. The three mammalian DNA methyltransferasess (DNMT1, DNMT3A, and DNMT3B) catalyze the addition of methyl groups to cytosines in DNA. While DNMT1 is a maintenance methyltransferase, DNMT3A and DNMT3B can carry out new methylations. There are also two splice protein isoforms produced from the ''DNMT3A'' gene: DNA methyltransferase proteins DNMT3A1 and DNMT3A2.
The splice isoform DNMT3A2 behaves like the product of a classical immediate-early gene and, for instance, it is robustly and transiently produced after neuronal activation. Where the DNA methyltransferase isoform DNMT3A2 binds and adds methyl groups to cytosines appears to be determined by histone post translational modifications.
On the other hand, neural activation causes degradation of DNMT3A1 accompanied by reduced methylation of at least one evaluated targeted promoter.
 One strand of the DNA, the ''template strand'' (or noncoding strand), is used as a template for RNA synthesis. As transcription proceeds, RNA polymerase traverses the template strand and uses base pairing complementarity with the DNA template to create an RNA copy (which elongates during the traversal). Although RNA polymerase traverses the template strand from 3' → 5', the coding (non-template) strand and newly formed RNA can also be used as reference points, so transcription can be described as occurring 5' → 3'. This produces an RNA molecule from 5' → 3', an exact copy of the coding strand (except that
One strand of the DNA, the ''template strand'' (or noncoding strand), is used as a template for RNA synthesis. As transcription proceeds, RNA polymerase traverses the template strand and uses base pairing complementarity with the DNA template to create an RNA copy (which elongates during the traversal). Although RNA polymerase traverses the template strand from 3' → 5', the coding (non-template) strand and newly formed RNA can also be used as reference points, so transcription can be described as occurring 5' → 3'. This produces an RNA molecule from 5' → 3', an exact copy of the coding strand (except that
 RNA polymerase plays a very crucial role in all steps including post-transcriptional changes in RNA.
RNA polymerase plays a very crucial role in all steps including post-transcriptional changes in RNA.
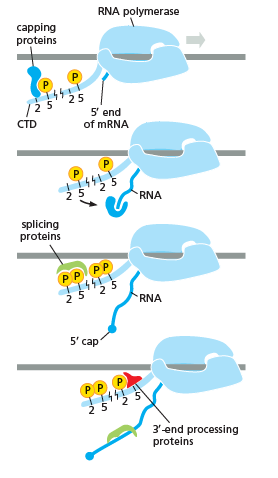 As shown in the image in the right it is evident that the CTD (C Terminal Domain) is a tail that changes its shape; this tail will be used as a carrier of splicing, capping and
As shown in the image in the right it is evident that the CTD (C Terminal Domain) is a tail that changes its shape; this tail will be used as a carrier of splicing, capping and
 Transcription can be measured and detected in a variety of ways:
* G-Less Cassette transcription assay: measures promoter strength
* Run-off transcription assay: identifies transcription start sites (TSS)
* Nuclear run-on assay: measures the relative abundance of newly formed transcripts
* KAS-seq: measures single-stranded DNA generated by RNA polymerases; can work with 1,000 cells.
* RNase protection assay and ChIP-Chip of RNAP: detect active transcription sites
* RT-PCR: measures the absolute abundance of total or nuclear RNA levels, which may however differ from transcription rates
* DNA microarrays: measures the relative abundance of the global total or nuclear RNA levels; however, these may differ from transcription rates
*
Transcription can be measured and detected in a variety of ways:
* G-Less Cassette transcription assay: measures promoter strength
* Run-off transcription assay: identifies transcription start sites (TSS)
* Nuclear run-on assay: measures the relative abundance of newly formed transcripts
* KAS-seq: measures single-stranded DNA generated by RNA polymerases; can work with 1,000 cells.
* RNase protection assay and ChIP-Chip of RNAP: detect active transcription sites
* RT-PCR: measures the absolute abundance of total or nuclear RNA levels, which may however differ from transcription rates
* DNA microarrays: measures the relative abundance of the global total or nuclear RNA levels; however, these may differ from transcription rates
*
 Some
Some
Interactive Java simulation of transcription initiation.
Fro
Center for Models of Life
at the Niels Bohr Institute.
From ttp://cmol.nbi.dk/ Center for Models of Life at the Niels Bohr Institute.
Virtual Cell Animation Collection, Introducing Transcription
{{DEFAULTSORT:Transcription (Genetics) Gene expression Molecular biology Cellular processes
DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
into RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
for the purpose of gene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
. Some segments of DNA are transcribed into RNA molecules that can encode protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s, called messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
(mRNA). Other segments of DNA are transcribed into RNA molecules called non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not Translation (genetics), translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally imp ...
s (ncRNAs).
Both DNA and RNA are nucleic acid
Nucleic acids are large biomolecules that are crucial in all cells and viruses. They are composed of nucleotides, which are the monomer components: a pentose, 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nuclei ...
s, which use base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s of nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
s as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase
In biochemistry, a polymerase is an enzyme (Enzyme Commission number, EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by ...
, which produces a complementary, antiparallel RNA strand called a primary transcript
A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNA ...
.
In virology
Virology is the Scientific method, scientific study of biological viruses. It is a subfield of microbiology that focuses on their detection, structure, classification and evolution, their methods of infection and exploitation of host (biology), ...
, the term transcription is used when referring to mRNA synthesis from a viral RNA molecule. The genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
of many RNA viruses is composed of negative-sense RNA which acts as a template for positive sense viral messenger RNA - a necessary step in the synthesis of viral proteins needed for viral replication
Viral replication is the formation of biological viruses during the infection process in the target host cells. Viruses must first get into the cell before viral replication can occur. Through the generation of abundant copies of its genome ...
. This process is catalyzed by a viral RNA dependent RNA polymerase.
Background
A DNA transcription unit encoding for a protein may contain both a ''coding sequence'', which will be translated into the protein, and ''regulatory sequences'', which direct and regulate the synthesis of that protein. The regulatory sequence before ( upstream from) the coding sequence is called the five prime untranslated regions (5'UTR); the sequence after ( downstream from) the coding sequence is called the three prime untranslated regions (3'UTR).Eldra P. Solomon, Linda R. Berg, Diana W. Martin. ''Biology, 8th Edition, International Student Edition''. Thomson Brooks/Cole. As opposed toDNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
, transcription results in an RNA complement that includes the nucleotide uracil
Uracil () (nucleoside#List of nucleosides and corresponding nucleobases, symbol U or Ura) is one of the four nucleotide bases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via ...
(U) in all instances where thymine
Thymine () (symbol T or Thy) is one of the four nucleotide bases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine ...
(T) would have occurred in a DNA complement.
Only one of the two DNA strands serves as a template for transcription. The antisense strand of DNA is read by RNA polymerase from the 3' end to the 5' end during transcription (3' → 5'). The complementary RNA is created in the opposite direction, in the 5' → 3' direction, matching the sequence of the sense strand except switching uracil for thymine. This directionality is because RNA polymerase can only add nucleotides to the 3' end of the growing mRNA chain. This use of only the 3' → 5' DNA strand eliminates the need for the Okazaki fragments that are seen in DNA replication. This also removes the need for an RNA primer to initiate RNA synthesis, as is the case in DNA replication.
The ''non''-template (sense) strand of DNA is called the coding strand, because its sequence is the same as the newly created RNA transcript (except for the substitution of uracil for thymine). This is the strand that is used by convention when presenting a DNA sequence.
Transcription has some proofreading mechanisms, but they are fewer and less effective than the controls for copying DNA. As a result, transcription has a lower copying fidelity than DNA replication.
Major steps
Transcription is divided into ''initiation'', ''promoter escape'', ''elongation,'' and ''termination''.Setting up for transcription
Enhancers, transcription factors, Mediator complex, and DNA loops in mammalian transcription
 Setting up for transcription in mammals is regulated by many cis-regulatory elements, including core promoter and promoter-proximal elements that are located near the transcription start sites of genes. Core promoters combined with general transcription factors are sufficient to direct transcription initiation, but generally have low basal activity. Other important cis-regulatory modules are localized in DNA regions that are distant from the transcription start sites. These include enhancers, silencers, insulators and tethering elements. Among this constellation of elements, enhancers and their associated transcription factors have a leading role in the initiation of gene transcription. An enhancer localized in a DNA region distant from the promoter of a gene can have a very large effect on gene transcription, with some genes undergoing up to 100-fold increased transcription due to an activated enhancer.
Enhancers are regions of the genome that are major gene-regulatory elements. Enhancers control cell-type-specific gene transcription programs, most often by looping through long distances to come in physical proximity with the promoters of their target genes. While there are hundreds of thousands of enhancer DNA regions, for a particular type of tissue only specific enhancers are brought into proximity with the promoters that they regulate. In a study of brain cortical neurons, 24,937 loops were found, bringing enhancers to their target promoters. Multiple enhancers, each often at tens or hundred of thousands of nucleotides distant from their target genes, loop to their target gene promoters and can coordinate with each other to control transcription of their common target gene.
The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene. The loop is stabilized by a dimer of a connector protein (e.g. dimer of CTCF or YY1), with one member of the dimer anchored to its binding motif on the enhancer and the other member anchored to its binding motif on the promoter (represented by the red zigzags in the illustration). Several cell function specific transcription factors (there are about 1,600 transcription factors in a human cell) generally bind to specific motifs on an enhancer and a small combination of these enhancer-bound transcription factors, when brought close to a promoter by a DNA loop, govern level of transcription of the target gene. Mediator (a complex usually consisting of about 26 proteins in an interacting structure) communicates regulatory signals from enhancer DNA-bound transcription factors directly to the RNA polymerase II (pol II) enzyme bound to the promoter.
Enhancers, when active, are generally transcribed from both strands of DNA with RNA polymerases acting in two different directions, producing two enhancer RNAs (eRNAs) as illustrated in the Figure. An inactive enhancer may be bound by an inactive transcription factor. Phosphorylation of the transcription factor may activate it and that activated transcription factor may then activate the enhancer to which it is bound (see small red star representing phosphorylation of transcription factor bound to enhancer in the illustration). An activated enhancer begins transcription of its RNA before activating transcription of messenger RNA from its target gene.
Setting up for transcription in mammals is regulated by many cis-regulatory elements, including core promoter and promoter-proximal elements that are located near the transcription start sites of genes. Core promoters combined with general transcription factors are sufficient to direct transcription initiation, but generally have low basal activity. Other important cis-regulatory modules are localized in DNA regions that are distant from the transcription start sites. These include enhancers, silencers, insulators and tethering elements. Among this constellation of elements, enhancers and their associated transcription factors have a leading role in the initiation of gene transcription. An enhancer localized in a DNA region distant from the promoter of a gene can have a very large effect on gene transcription, with some genes undergoing up to 100-fold increased transcription due to an activated enhancer.
Enhancers are regions of the genome that are major gene-regulatory elements. Enhancers control cell-type-specific gene transcription programs, most often by looping through long distances to come in physical proximity with the promoters of their target genes. While there are hundreds of thousands of enhancer DNA regions, for a particular type of tissue only specific enhancers are brought into proximity with the promoters that they regulate. In a study of brain cortical neurons, 24,937 loops were found, bringing enhancers to their target promoters. Multiple enhancers, each often at tens or hundred of thousands of nucleotides distant from their target genes, loop to their target gene promoters and can coordinate with each other to control transcription of their common target gene.
The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene. The loop is stabilized by a dimer of a connector protein (e.g. dimer of CTCF or YY1), with one member of the dimer anchored to its binding motif on the enhancer and the other member anchored to its binding motif on the promoter (represented by the red zigzags in the illustration). Several cell function specific transcription factors (there are about 1,600 transcription factors in a human cell) generally bind to specific motifs on an enhancer and a small combination of these enhancer-bound transcription factors, when brought close to a promoter by a DNA loop, govern level of transcription of the target gene. Mediator (a complex usually consisting of about 26 proteins in an interacting structure) communicates regulatory signals from enhancer DNA-bound transcription factors directly to the RNA polymerase II (pol II) enzyme bound to the promoter.
Enhancers, when active, are generally transcribed from both strands of DNA with RNA polymerases acting in two different directions, producing two enhancer RNAs (eRNAs) as illustrated in the Figure. An inactive enhancer may be bound by an inactive transcription factor. Phosphorylation of the transcription factor may activate it and that activated transcription factor may then activate the enhancer to which it is bound (see small red star representing phosphorylation of transcription factor bound to enhancer in the illustration). An activated enhancer begins transcription of its RNA before activating transcription of messenger RNA from its target gene.
CpG island methylation and demethylation
DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
base cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
(see Figure). 5-mC is an epigenetic marker found predominantly within CpG sites. About 28 million CpG dinucleotides occur in the human genome. In most tissues of mammals, on average, 70% to 80% of CpG cytosines are methylated (forming 5-methylCpG or 5-mCpG). However, unmethylated cytosines within 5'cytosine-guanine 3' sequences often occur in groups, called CpG islands, at active promoters. About 60% of promoter sequences have a CpG island while only about 6% of enhancer sequences have a CpG island. CpG islands constitute regulatory sequences, since if CpG islands are methylated in the promoter of a gene this can reduce or silence gene transcription.
DNA methylation regulates gene transcription through interaction with methyl binding domain (MBD) proteins, such as MeCP2, MBD1 and MBD2. These MBD proteins bind most strongly to highly methylated CpG islands. These MBD proteins have both a methyl-CpG-binding domain as well as a transcription repression domain. They bind to methylated DNA and guide or direct protein complexes with chromatin remodeling and/or histone modifying activity to methylated CpG islands. MBD proteins generally repress local chromatin such as by catalyzing the introduction of repressive histone marks, or creating an overall repressive chromatin environment through nucleosome remodeling and chromatin reorganization.
 As noted in the previous section, transcription factors are proteins that bind to specific DNA sequences in order to regulate the expression of a gene. The binding sequence for a transcription factor in DNA is usually about 10 or 11 nucleotides long. As summarized in 2009, Vaquerizas et al. indicated there are approximately 1,400 different transcription factors encoded in the human genome by genes that constitute about 6% of all human protein encoding genes. About 94% of transcription factor binding sites (TFBSs) that are associated with signal-responsive genes occur in enhancers while only about 6% of such TFBSs occur in promoters.
EGR1 protein is a particular transcription factor that is important for regulation of methylation of CpG islands. An EGR1 transcription factor binding site is frequently located in enhancer or promoter sequences. There are about 12,000 binding sites for EGR1 in the mammalian genome and about half of EGR1 binding sites are located in promoters and half in enhancers. The binding of EGR1 to its target DNA binding site is insensitive to cytosine methylation in the DNA.
While only small amounts of EGR1 transcription factor protein are detectable in cells that are un-stimulated, translation of the ''EGR1'' gene into protein at one hour after stimulation is drastically elevated. Production of EGR1 transcription factor proteins, in various types of cells, can be stimulated by growth factors, neurotransmitters, hormones, stress and injury. In the brain, when neurons are activated, EGR1 proteins are up-regulated and they bind to (recruit) the pre-existing TET1 enzymes that are produced in high amounts in neurons. TET enzymes can catalyse demethylation of 5-methylcytosine. When EGR1 transcription factors bring TET1 enzymes to EGR1 binding sites in promoters, the TET enzymes can demethylate the methylated CpG islands at those promoters. Upon demethylation, these promoters can then initiate transcription of their target genes. Hundreds of genes in neurons are differentially expressed after neuron activation through EGR1 recruitment of TET1 to methylated regulatory sequences in their promoters.
The methylation of promoters is also altered in response to signals. The three mammalian DNA methyltransferasess (DNMT1, DNMT3A, and DNMT3B) catalyze the addition of methyl groups to cytosines in DNA. While DNMT1 is a maintenance methyltransferase, DNMT3A and DNMT3B can carry out new methylations. There are also two splice protein isoforms produced from the ''DNMT3A'' gene: DNA methyltransferase proteins DNMT3A1 and DNMT3A2.
The splice isoform DNMT3A2 behaves like the product of a classical immediate-early gene and, for instance, it is robustly and transiently produced after neuronal activation. Where the DNA methyltransferase isoform DNMT3A2 binds and adds methyl groups to cytosines appears to be determined by histone post translational modifications.
On the other hand, neural activation causes degradation of DNMT3A1 accompanied by reduced methylation of at least one evaluated targeted promoter.
As noted in the previous section, transcription factors are proteins that bind to specific DNA sequences in order to regulate the expression of a gene. The binding sequence for a transcription factor in DNA is usually about 10 or 11 nucleotides long. As summarized in 2009, Vaquerizas et al. indicated there are approximately 1,400 different transcription factors encoded in the human genome by genes that constitute about 6% of all human protein encoding genes. About 94% of transcription factor binding sites (TFBSs) that are associated with signal-responsive genes occur in enhancers while only about 6% of such TFBSs occur in promoters.
EGR1 protein is a particular transcription factor that is important for regulation of methylation of CpG islands. An EGR1 transcription factor binding site is frequently located in enhancer or promoter sequences. There are about 12,000 binding sites for EGR1 in the mammalian genome and about half of EGR1 binding sites are located in promoters and half in enhancers. The binding of EGR1 to its target DNA binding site is insensitive to cytosine methylation in the DNA.
While only small amounts of EGR1 transcription factor protein are detectable in cells that are un-stimulated, translation of the ''EGR1'' gene into protein at one hour after stimulation is drastically elevated. Production of EGR1 transcription factor proteins, in various types of cells, can be stimulated by growth factors, neurotransmitters, hormones, stress and injury. In the brain, when neurons are activated, EGR1 proteins are up-regulated and they bind to (recruit) the pre-existing TET1 enzymes that are produced in high amounts in neurons. TET enzymes can catalyse demethylation of 5-methylcytosine. When EGR1 transcription factors bring TET1 enzymes to EGR1 binding sites in promoters, the TET enzymes can demethylate the methylated CpG islands at those promoters. Upon demethylation, these promoters can then initiate transcription of their target genes. Hundreds of genes in neurons are differentially expressed after neuron activation through EGR1 recruitment of TET1 to methylated regulatory sequences in their promoters.
The methylation of promoters is also altered in response to signals. The three mammalian DNA methyltransferasess (DNMT1, DNMT3A, and DNMT3B) catalyze the addition of methyl groups to cytosines in DNA. While DNMT1 is a maintenance methyltransferase, DNMT3A and DNMT3B can carry out new methylations. There are also two splice protein isoforms produced from the ''DNMT3A'' gene: DNA methyltransferase proteins DNMT3A1 and DNMT3A2.
The splice isoform DNMT3A2 behaves like the product of a classical immediate-early gene and, for instance, it is robustly and transiently produced after neuronal activation. Where the DNA methyltransferase isoform DNMT3A2 binds and adds methyl groups to cytosines appears to be determined by histone post translational modifications.
On the other hand, neural activation causes degradation of DNMT3A1 accompanied by reduced methylation of at least one evaluated targeted promoter.
Initiation
Transcription begins with the RNA polymerase and one or more general transcription factors binding to a DNA promoter sequence to form an RNA polymerase-promoter closed complex. In the closed complex, the promoter DNA is still fully double-stranded. RNA polymerase, assisted by one or more general transcription factors, then unwinds approximately 14 base pairs of DNA to form an RNA polymerase-promoter open complex. In the open complex, the promoter DNA is partly unwound and single-stranded. The exposed, single-stranded DNA is referred to as the " transcription bubble". RNA polymerase, assisted by one or more general transcription factors, then selects a transcription start site in the transcription bubble, binds to an initiating NTP and an extending NTP (or a short RNA primer and an extending NTP) complementary to the transcription start site sequence, and catalyzes bond formation to yield an initial RNA product. Inbacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
, RNA polymerase holoenzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
consists of five subunits: 2 α subunits, 1 β subunit, 1 β' subunit, and 1 ω subunit. In bacteria, there is one general RNA transcription factor known as a sigma factor
A sigma factor (σ factor or specificity factor) is a protein needed for initiation of Transcription (biology), transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to g ...
. RNA polymerase core enzyme binds to the bacterial general transcription (sigma) factor to form RNA polymerase holoenzyme and then binds to a promoter.
(RNA polymerase is called a holoenzyme when sigma subunit is attached to the core enzyme which is consist of 2 α subunits, 1 β subunit, 1 β' subunit only). Unlike eukaryotes, the initiating nucleotide of nascent bacterial mRNA is not capped with a modified guanine nucleotide. The initiating nucleotide of bacterial transcripts bears a 5′ triphosphate (5′-PPP), which can be used for genome-wide mapping of transcription initiation sites.
In archaea
Archaea ( ) is a Domain (biology), domain of organisms. Traditionally, Archaea only included its Prokaryote, prokaryotic members, but this has since been found to be paraphyletic, as eukaryotes are known to have evolved from archaea. Even thou ...
and eukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of ...
, RNA polymerase contains subunits homologous to each of the five RNA polymerase subunits in bacteria and also contains additional subunits. In archaea and eukaryotes, the functions of the bacterial general transcription factor sigma are performed by multiple general transcription factors that work together. In archaea, there are three general transcription factors: TBP, TFB, and TFE. In eukaryotes, in RNA polymerase II
RNA polymerase II (RNAP II and Pol II) is a Protein complex, multiprotein complex that Transcription (biology), transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNA pol ...
-dependent transcription, there are six general transcription factors: TFIIA, TFIIB (an ortholog of archaeal TFB), TFIID (a multisubunit factor in which the key subunit, TBP, is an ortholog of archaeal TBP), TFIIE (an ortholog of archaeal TFE), TFIIF, and TFIIH. The TFIID is the first component to bind to DNA due to binding of TBP, while TFIIH is the last component to be recruited. In archaea and eukaryotes, the RNA polymerase-promoter closed complex is usually referred to as the " preinitiation complex".
Transcription initiation is regulated by additional proteins, known as activators and repressor
In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the ...
s, and, in some cases, associated coactivators or corepressors, which modulate formation and function of the transcription initiation complex.
Promoter escape
After the first bond is synthesized, the RNA polymerase must escape the promoter. During this time there is a tendency to release the RNA transcript and produce truncated transcripts. This is called abortive initiation, and is common for both eukaryotes and prokaryotes. Abortive initiation continues to occur until an RNA product of a threshold length of approximately 10 nucleotides is synthesized, at which point promoter escape occurs and a transcription elongation complex is formed. Mechanistically, promoter escape occurs through DNA scrunching, providing the energy needed to break interactions between RNA polymerase holoenzyme and the promoter. In bacteria, it was historically thought that thesigma factor
A sigma factor (σ factor or specificity factor) is a protein needed for initiation of Transcription (biology), transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to g ...
is definitely released after promoter clearance occurs. This theory had been known as the ''obligate release model.'' However, later data showed that upon and following promoter clearance, the sigma factor is released according to a stochastic model known as the ''stochastic release model''.
In eukaryotes, at an RNA polymerase II-dependent promoter, upon promoter clearance, TFIIH phosphorylates serine 5 on the carboxy terminal domain of RNA polymerase II, leading to the recruitment of capping enzyme (CE). The exact mechanism of how CE induces promoter clearance in eukaryotes is not yet known.
Elongation
thymine
Thymine () (symbol T or Thy) is one of the four nucleotide bases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine ...
s are replaced with uracil
Uracil () (nucleoside#List of nucleosides and corresponding nucleobases, symbol U or Ura) is one of the four nucleotide bases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via ...
s, and the nucleotides are composed of a ribose (5-carbon) sugar whereas DNA has deoxyribose (one fewer oxygen atom) in its sugar-phosphate backbone).
mRNA transcription can involve multiple RNA polymerases on a single DNA template and multiple rounds of transcription (amplification of particular mRNA), so many mRNA molecules can be rapidly produced from a single copy of a gene. The characteristic elongation rates in prokaryotes and eukaryotes are about 10–100 nts/sec. In eukaryotes, however, nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
s act as major barriers to transcribing polymerases during transcription elongation. In these organisms, the pausing induced by nucleosomes can be regulated by transcription elongation factors such as TFIIS.
Elongation also involves a proofreading mechanism that can replace incorrectly incorporated bases. In eukaryotes, this may correspond with short pauses during transcription that allow appropriate RNA editing factors to bind. These pauses may be intrinsic to the RNA polymerase or due to chromatin structure.
Double-strand breaks in actively transcribed regions of DNA are repaired by homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in Cell (biology), cellular organi ...
during the S and G2 phases of the cell cycle
The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell (biology), cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA (DNA re ...
. Since transcription enhances the accessibility of DNA to exogenous chemicals and internal metabolites that can cause recombinogenic lesions, homologous recombination of a particular DNA sequence may be strongly stimulated by transcription.
Termination
Bacteria use two different strategies for transcription termination – Rho-independent termination and Rho-dependent termination. In Rho-independent transcription termination, RNA transcription stops when the newly synthesized RNA molecule forms a G-C-rich hairpin loop followed by a run of Us. When the hairpin forms, the mechanical stress breaks the weak rU-dA bonds, now filling the DNA–RNA hybrid. This pulls the poly-U transcript out of the active site of the RNA polymerase, terminating transcription. In Rho-dependent termination,Rho
Rho (; uppercase Ρ, lowercase ρ or ; or ) is the seventeenth letter of the Greek alphabet. In the system of Greek numerals it has a value of 100. It is derived from Phoenician alphabet, Phoenician letter resh . Its uppercase form uses the same ...
, a protein factor, destabilizes the interaction between the template and the mRNA, thus releasing the newly synthesized mRNA from the elongation complex.
Transcription termination in eukaryotes is less well understood than in bacteria, but involves cleavage of the new transcript followed by template-independent addition of adenines at its new 3' end, in a process called polyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In euka ...
.
Beyond termination by a terminator sequences (which is a part of a gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
), transcription may also need to be terminated when it encounters conditions such as DNA damage or an active replication fork. In bacteria, the Mfd ATPase can remove a RNA polymerase stalled at a lesion by prying open its clamp. It also recruits nucleotide excision repair machinery to repair the lesion. Mfd is proposed to also resolve conflicts between DNA replication and transcription. In eukayrotes, ATPase TTF2 helps to suppress the action of RNAP I and II during mitosis
Mitosis () is a part of the cell cycle in eukaryote, eukaryotic cells in which replicated chromosomes are separated into two new Cell nucleus, nuclei. Cell division by mitosis is an equational division which gives rise to genetically identic ...
, preventing errors in chromosomal segregation. In archaea, the Eta ATPase is proposed to play a similar role.
Transcription increases susceptibility to DNA damage
Genome damage occurs with a high frequency, estimated to range between tens and hundreds of thousands of DNA damages arising in each cell every day. The process of transcription is a major source of DNA damage, due to the formation of single-strand DNA intermediates that are vulnerable to damage. The regulation of transcription by processes using base excision repair and/or topoisomerases to cut and remodel the genome also increases the vulnerability of DNA to damage.Role of RNA polymerase in post-transcriptional changes in RNA
 RNA polymerase plays a very crucial role in all steps including post-transcriptional changes in RNA.
RNA polymerase plays a very crucial role in all steps including post-transcriptional changes in RNA.
 As shown in the image in the right it is evident that the CTD (C Terminal Domain) is a tail that changes its shape; this tail will be used as a carrier of splicing, capping and
As shown in the image in the right it is evident that the CTD (C Terminal Domain) is a tail that changes its shape; this tail will be used as a carrier of splicing, capping and polyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In euka ...
, as shown in the image on the left.
Inhibitors
Transcription inhibitors can be used asantibiotic
An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting pathogenic bacteria, bacterial infections, and antibiotic medications are widely used in the therapy ...
s against, for example, pathogenic bacteria
Pathogenic bacteria are bacteria that can cause disease. This article focuses on the bacteria that are pathogenic to humans. Most species of bacteria are harmless and many are Probiotic, beneficial but others can cause infectious diseases. The nu ...
( antibacterials) and fungi
A fungus (: fungi , , , or ; or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and mold (fungus), molds, as well as the more familiar mushrooms. These organisms are classified as one ...
(antifungals
An antifungal medication, also known as an antimycotic medication, is a pharmaceutical fungicide or fungistatic used to treat and prevent mycosis such as athlete's foot, ringworm, candidiasis (thrush), serious systemic infections such as ...
). An example of such an antibacterial is rifampicin, which inhibits bacterial transcription of DNA into mRNA by inhibiting DNA-dependent RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
by binding its beta-subunit, while 8-hydroxyquinoline is an antifungal transcription inhibitor. The effects of histone methylation may also work to inhibit the action of transcription. Potent, bioactive natural products like triptolide that inhibit mammalian transcription via inhibition of the XPB subunit of the general transcription factor TFIIH has been recently reported as a glucose conjugate for targeting hypoxic cancer cells with increased glucose transporter production.
Endogenous inhibitors
In vertebrates, the majority of gene promoters contain a CpG island with numerous CpG sites. When many of a gene's promoter CpG sites are methylated the gene becomes inhibited (silenced). Colorectal cancers typically have 3 to 6 driver mutations and 33 to 66 hitchhiker or passenger mutations. However, transcriptional inhibition (silencing) may be of more importance than mutation in causing progression to cancer. For example, in colorectal cancers about 600 to 800 genes are transcriptionally inhibited by CpG island methylation (see regulation of transcription in cancer). Transcriptional repression in cancer can also occur by other epigenetic mechanisms, such as altered production of microRNAs. In breast cancer, transcriptional repression of BRCA1 may occur more frequently by over-produced microRNA-182 than by hypermethylation of the BRCA1 promoter (see Low expression of BRCA1 in breast and ovarian cancers).Transcription factories
Active transcription units are clustered in the nucleus, in discrete sites called transcription factories or euchromatin. Such sites can be visualized by allowing engaged polymerases to extend their transcripts in tagged precursors (Br-UTP or Br-U) and immuno-labeling the tagged nascent RNA. Transcription factories can also be localized using fluorescence in situ hybridization or marked by antibodies directed against polymerases. There are ≈10,000 factories in the nucleoplasm of a HeLa cell, among which are ≈8,000 polymerase II factories and ≈2,000 polymerase III factories. Each polymerase II factory contains ≈8 polymerases. As most active transcription units are associated with only one polymerase, each factory usually contains ≈8 different transcription units. These units might be associated through promoters and/or enhancers, with loops forming a "cloud" around the factor.History
A molecule that allows the genetic material to be realized as a protein was first hypothesized byFrançois Jacob
François Jacob (; 17 June 1920 – 19 April 2013) was a French biologist who, together with Jacques Monod, originated the idea that control of enzyme levels in all cells occurs through regulation of transcription. He shared the 1965 Nobel ...
and Jacques Monod
Jacques Lucien Monod (; 9 February 1910 – 31 May 1976) was a French biochemist who won the Nobel Prize in Physiology or Medicine in 1965, sharing it with François Jacob and André Lwoff "for their discoveries concerning genetic control of e ...
. Severo Ochoa won a Nobel Prize in Physiology or Medicine
The Nobel Prize in Physiology or Medicine () is awarded yearly by the Nobel Assembly at the Karolinska Institute for outstanding discoveries in physiology or medicine. The Nobel Prize is not a single prize, but five separate prizes that, acco ...
in 1959 for developing a process for synthesizing RNA ''in vitro
''In vitro'' (meaning ''in glass'', or ''in the glass'') Research, studies are performed with Cell (biology), cells or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in ...
'' with polynucleotide phosphorylase, which was useful for cracking the genetic code
Genetic code is a set of rules used by living cell (biology), cells to Translation (biology), translate information encoded within genetic material (DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished ...
. RNA synthesis by RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
was established ''in vitro'' by several laboratories by 1965; however, the RNA synthesized by these enzymes had properties that suggested the existence of an additional factor needed to terminate transcription correctly.
Roger D. Kornberg won the 2006 Nobel Prize in Chemistry "for his studies of the molecular basis of eukaryotic transcription".
Measuring and detecting
 Transcription can be measured and detected in a variety of ways:
* G-Less Cassette transcription assay: measures promoter strength
* Run-off transcription assay: identifies transcription start sites (TSS)
* Nuclear run-on assay: measures the relative abundance of newly formed transcripts
* KAS-seq: measures single-stranded DNA generated by RNA polymerases; can work with 1,000 cells.
* RNase protection assay and ChIP-Chip of RNAP: detect active transcription sites
* RT-PCR: measures the absolute abundance of total or nuclear RNA levels, which may however differ from transcription rates
* DNA microarrays: measures the relative abundance of the global total or nuclear RNA levels; however, these may differ from transcription rates
*
Transcription can be measured and detected in a variety of ways:
* G-Less Cassette transcription assay: measures promoter strength
* Run-off transcription assay: identifies transcription start sites (TSS)
* Nuclear run-on assay: measures the relative abundance of newly formed transcripts
* KAS-seq: measures single-stranded DNA generated by RNA polymerases; can work with 1,000 cells.
* RNase protection assay and ChIP-Chip of RNAP: detect active transcription sites
* RT-PCR: measures the absolute abundance of total or nuclear RNA levels, which may however differ from transcription rates
* DNA microarrays: measures the relative abundance of the global total or nuclear RNA levels; however, these may differ from transcription rates
* In situ hybridization
''In situ'' hybridization (ISH) is a type of Hybridisation (molecular biology), hybridization that uses a labeled complementary DNA, RNA or modified nucleic acid strand (i.e., a Hybridization probe, probe) to localize a specific DNA or RNA seq ...
: detects the presence of a transcript
* MS2 tagging: by incorporating RNA stem loops, such as MS2, into a gene, these become incorporated into newly synthesized RNA. The stem loops can then be detected using a fusion of GFP and the MS2 coat protein, which has a high affinity, sequence-specific interaction with the MS2 stem loops. The recruitment of GFP to the site of transcription is visualized as a single fluorescent spot. This new approach has revealed that transcription occurs in discontinuous bursts, or pulses (see Transcriptional bursting). With the notable exception of in situ techniques, most other methods provide cell population averages, and are not capable of detecting this fundamental property of genes.
* Northern blot: the traditional method, and until the advent of RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also k ...
, the most quantitative
* RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also k ...
: applies next-generation sequencing techniques to sequence whole transcriptomes, which allows the measurement of relative abundance of RNA, as well as the detection of additional variations such as fusion genes, post-transcriptional edits and novel splice sites
* Single cell RNA-Seq: amplifies and reads partial transcriptomes from isolated cells, allowing for detailed analyses of RNA in tissues, embryos, and cancers
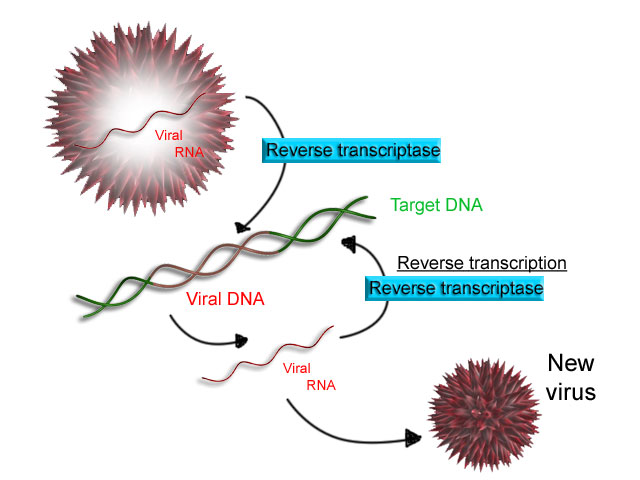
Reverse transcription
 Some
Some viruses
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Viruses are found in almo ...
(such as HIV, the cause of AIDS
The HIV, human immunodeficiency virus (HIV) is a retrovirus that attacks the immune system. Without treatment, it can lead to a spectrum of conditions including acquired immunodeficiency syndrome (AIDS). It is a Preventive healthcare, pr ...
), have the ability to transcribe RNA into DNA. HIV has an RNA genome that is ''reverse transcribed'' into DNA. The resulting DNA can be merged with the DNA genome of the host cell. The main enzyme responsible for synthesis of DNA from an RNA template is called reverse transcriptase
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobi ...
.
In the case of HIV, reverse transcriptase is responsible for synthesizing a complementary DNA strand (cDNA) to the viral RNA genome. The enzyme ribonuclease H then digests the RNA strand, and reverse transcriptase synthesises a complementary strand of DNA to form a double helix DNA structure (cDNA). The cDNA is integrated into the host cell's genome by the enzyme integrase
Retroviral integrase (IN) is an enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme ...
, which causes the host cell to generate viral proteins that reassemble into new viral particles. In HIV, subsequent to this, the host cell undergoes programmed cell death, or apoptosis
Apoptosis (from ) is a form of programmed cell death that occurs in multicellular organisms and in some eukaryotic, single-celled microorganisms such as yeast. Biochemistry, Biochemical events lead to characteristic cell changes (Morphology (biol ...
, of T cell
T cells (also known as T lymphocytes) are an important part of the immune system and play a central role in the adaptive immune response. T cells can be distinguished from other lymphocytes by the presence of a T-cell receptor (TCR) on their cell ...
s. However, in other retroviruses, the host cell remains intact as the virus buds out of the cell.
Some eukaryotic cells contain an enzyme with reverse transcription activity called telomerase. Telomerase carries an RNA template from which it synthesizes a telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes (see #Sequences, Sequences). Telomeres are a widespread genetic feature most commonly found in eukaryotes. In ...
, a repeating sequence of DNA, to the end of linear chromosomes. It is important because every time a linear chromosome is duplicated, it is shortened. With the telomere at the ends of chromosomes, the shortening eliminates some of the non-essential, repeated sequence, rather than the protein-encoding DNA sequence farther away from the chromosome end.
Telomerase is often activated in cancer cells to enable cancer cells to duplicate their genomes indefinitely without losing important protein-coding DNA sequence. Activation of telomerase could be part of the process that allows cancer cells to become immortal. The immortalizing factor of cancer via telomere lengthening due to telomerase has been proven to occur in 90% of all carcinogenic tumors ''in vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, an ...
'' with the remaining 10% using an alternative telomere maintenance route called ALT or Alternative Lengthening of Telomeres.
See also
*Life
Life, also known as biota, refers to matter that has biological processes, such as Cell signaling, signaling and self-sustaining processes. It is defined descriptively by the capacity for homeostasis, Structure#Biological, organisation, met ...
* Cell (biology)
The cell is the basic structural and functional unit of all life, forms of life. Every cell consists of cytoplasm enclosed within a Cell membrane, membrane; many cells contain organelles, each with a specific function. The term comes from the ...
* Cell division
Cell division is the process by which a parent cell (biology), cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukar ...
* DBTSS
* Gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
* Gene regulation
* Epigenetics
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
* Genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
* Gene regulation
* Long non-coding RNA
* Missense mRNA
* Splicing – process of removing intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gen ...
s from precursor messenger RNA (pre-mRNA
A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by Transcription (genetics), transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcript ...
) to make messenger RNA (mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
)
* Transcriptomics
* Translation (biology)
In biology, translation is the process in living Cell (biology), cells in which proteins are produced using RNA molecules as templates. The generated protein is a sequence of amino acids. This sequence is determined by the sequence of nucleotide ...
Notes
References
External links
Interactive Java simulation of transcription initiation.
Fro
Center for Models of Life
at the Niels Bohr Institute.
From ttp://cmol.nbi.dk/ Center for Models of Life at the Niels Bohr Institute.
Virtual Cell Animation Collection, Introducing Transcription
{{DEFAULTSORT:Transcription (Genetics) Gene expression Molecular biology Cellular processes