DNA demethylation on:
[Wikipedia]
[Google]
[Amazon]
 For
For

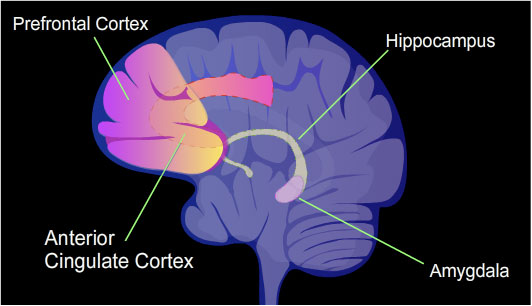 Learning and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature. Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time. Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory. At 24 hours after training, 9.17% of the genes in the genomes of rat hippocampus neurons were found to be differentially methylated. This included more than 2,000 differentially methylated genes at 24 hours after training, with over 500 genes being demethylated. Similar results to that in the rat hippocampus were also obtained in mice with contextual fear conditioning.
The hippocampus region of the brain is where contextual fear memories are first stored (see figure of the brain, this section), but this storage is transient and does not remain in the hippocampus. In rats contextual fear conditioning is abolished when the hippocampus is subjected to hippocampectomy just one day after conditioning, but rats retain a considerable amount of contextual fear when hippocampectomy is delayed by four weeks. In mice, examined at 4 weeks after conditioning, the hippocampus methylations and demethylations were reversed (the hippocampus is needed to form memories but memories are not stored there) while substantial differential CpG methylation and demethylation occurred in cortical neurons during memory maintenance. There were 1,223 differentially methylated genes in the anterior cingulate cortex of mice four weeks after contextual fear conditioning. Thus, while there were many methylations in the hippocampus shortly after memory was formed, all these hippocampus methylations were demethylated as soon as four weeks later.
Learning and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature. Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time. Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory. At 24 hours after training, 9.17% of the genes in the genomes of rat hippocampus neurons were found to be differentially methylated. This included more than 2,000 differentially methylated genes at 24 hours after training, with over 500 genes being demethylated. Similar results to that in the rat hippocampus were also obtained in mice with contextual fear conditioning.
The hippocampus region of the brain is where contextual fear memories are first stored (see figure of the brain, this section), but this storage is transient and does not remain in the hippocampus. In rats contextual fear conditioning is abolished when the hippocampus is subjected to hippocampectomy just one day after conditioning, but rats retain a considerable amount of contextual fear when hippocampectomy is delayed by four weeks. In mice, examined at 4 weeks after conditioning, the hippocampus methylations and demethylations were reversed (the hippocampus is needed to form memories but memories are not stored there) while substantial differential CpG methylation and demethylation occurred in cortical neurons during memory maintenance. There were 1,223 differentially methylated genes in the anterior cingulate cortex of mice four weeks after contextual fear conditioning. Thus, while there were many methylations in the hippocampus shortly after memory was formed, all these hippocampus methylations were demethylated as soon as four weeks later.
 Demethylation of 5-methylcytosine to generate 5-hydroxymethylcytosine (5hmC) very often initially involves oxidation of 5mC (see Figure in this section) by ten-eleven translocation methylcytosine dioxygenases (
Demethylation of 5-methylcytosine to generate 5-hydroxymethylcytosine (5hmC) very often initially involves oxidation of 5mC (see Figure in this section) by ten-eleven translocation methylcytosine dioxygenases (
 For a TET enzyme to initiate demethylation it must first be recruited to a methylated
For a TET enzyme to initiate demethylation it must first be recruited to a methylated
 The third stage of DNA demethylation is removal of the intermediate products of demethylation generated by a TET enzyme by
The third stage of DNA demethylation is removal of the intermediate products of demethylation generated by a TET enzyme by
molecular biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physi ...
in mammals, DNA demethylation Demethylation is the chemical process resulting in the removal of a methyl group (CH3) from a molecule. A common way of demethylation is the replacement of a methyl group by a hydrogen atom, resulting in a net loss of one carbon and two hydrogen at ...
causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine
Cytosine () ( symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an ...
(C) (see figure of 5mC and C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequence or, in replicating cells, by preventing addition of methyl groups to DNA so that the replicated DNA will largely have cytosine in the DNA sequence (5mC will be diluted out).
Methylated cytosine
Cytosine () ( symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an ...
is frequently present in the linear DNA sequence where a cytosine is followed by a guanine
Guanine () ( symbol G or Gua) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine (uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside is c ...
in a 5' → 3' direction (a CpG site
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG isl ...
). In mammals, DNA methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl m ...
s (which add methyl group
In organic chemistry, a methyl group is an alkyl derived from methane, containing one carbon atom bonded to three hydrogen atoms, having chemical formula . In formulas, the group is often abbreviated as Me. This hydrocarbon group occurs in ma ...
s to DNA bases) exhibit a strong sequence preference for cytosines at CpG site
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG isl ...
s. There appear to be more than 20 million CpG dinucleotides in the human genome (see genomic distribution). In mammals, on average, 70% to 80% of CpG cytosines are methylated, though the level of methylation varies with different tissues. Methylated cytosines often occur in groups or CpG islands within the promoter regions of genes, where such methylation may reduce or silence gene expression (see gene expression). Methylated cytosines in the gene body, however, are positively correlated with expression.
Almost 100% DNA demethylation occurs by a combination of passive dilution and active enzymatic removal during the reprogramming
In biology, reprogramming refers to erasure and remodeling of epigenetic marks, such as DNA methylation, during mammalian development or in cell culture. Such control is also often associated with alternative covalent modifications of histones.
...
that occurs in early embryogenesis and in gametogenesis
Gametogenesis is a biological process by which diploid or haploid precursor cells undergo cell division and differentiation to form mature haploid gametes. Depending on the biological life cycle of the organism, gametogenesis occurs by meiotic di ...
. Another large demethylation, of about 3% of all genes, can occur by active demethylation in neurons during formation of a strong memory. After surgery, demethylations are found in peripheral blood mononuclear cells at sites annotated to immune system genes. Demethylations also occur during the formation of cancers. During global DNA hypomethylation of tumor genomes, there is a minor to moderate reduction of the number of methylated cytosines (5mC) amounting to a loss of about 5% to 20% on average of the 5mC bases.
Embryonic development

Early embryonic development
The mouse spermgenome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding g ...
is 80–90% methylated
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These ...
at its CpG site
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG isl ...
s in DNA, amounting to about 20 million methylated sites. After fertilization
Fertilisation or fertilization (see spelling differences), also known as generative fertilisation, syngamy and impregnation, is the fusion of gametes to give rise to a new individual organism or offspring and initiate its development. Proce ...
, the paternal chromosome is almost completely demethylated in six hours by an active process, before DNA replication (blue line in Figure).
Demethylation of the maternal genome occurs by a different process. In the mature oocyte
An oocyte (, ), oöcyte, or ovocyte is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in a female fetus in the ovary during female gametogenesis. The femal ...
, about 40% of its CpG sites in DNA are methylated. While somatic cells of mammals have three main DNA methyltransferases (which add methyl groups to cytosines at CpG sites), DNMT1
DNA (cytosine-5)-methyltransferase 1 is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. In humans, it is encoded by the ''DNMT1'' gene. DNMT1 forms part of the family of ...
, DNMT3A
DNA (cytosine-5)-methyltransferase 3A is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. The enzyme is encoded in humans by the DNMT3A gene.
This enzyme is responsible f ...
, and DNMT3B
DNA (cytosine-5)-methyltransferase 3 beta, is an enzyme that in humans in encoded by the DNMT3B gene. Mutation in this gene are associated with immunodeficiency, centromere instability and facial anomalies syndrome.
Function
CpG methylation i ...
, in the pre-implantation embryo up to the blastocyst stage (see Figure), the only methyltransferase present is an isoform of DNMT1 designated DNMT1o. DNMT1o has an alternative oocyte-specific promoter and first exon (exon 1o) located 5' of the somatic and spermatocyte promoters. As reviewed by Howell et al., DNMT1o is sequestered in the cytoplasm of mature oocytes and in 2-cell and 4-cell embryos, but at the 8-cell stage is only present in the nucleus. At the 16 cell stage (the morula
A morula (Latin, ''morus'': mulberry) is an early-stage embryo consisting of a solid ball of cells called blastomeres, contained in mammals, and other animals within the zona pellucida shell. The blastomeres are the daughter cells of the zygot ...
) DNMT1o is again found only in the cytoplasm. It appears that demethylation of the maternal chromosomes largely takes place by blockage of the methylating enzyme DNMT1o from entering the nucleus except briefly at the 8 cell stage. The maternal-origin DNA thus undergoes passive demethylation by dilution of the methylated maternal DNA during replication (red line in Figure). The morula
A morula (Latin, ''morus'': mulberry) is an early-stage embryo consisting of a solid ball of cells called blastomeres, contained in mammals, and other animals within the zona pellucida shell. The blastomeres are the daughter cells of the zygot ...
(at the 16 cell stage), has only a small amount of DNA methylation (black line in Figure).
DNMT3b begins to be expressed in the blastocyst. Methylation begins to increase at 3.5 days after fertilization in the blastocyst, and a large wave of methylation then occurs on days 4.5 to 5.5 in the epiblast, going from 12% to 62% methylation, and reaching maximum level after implantation in the uterus. By day seven after fertilization, the newly formed primordial germ cells (PGC) in the implanted embryo segregate from the remaining somatic cells. At this point the PGCs have about the same level of methylation as the somatic cells.
Gametogenesis
The newly formed primordial germ cells (PGC) in the implanted embryo devolve from the somatic cells. At this point the PGCs have high levels of methylation. These cells migrate from the epiblast toward thegonadal ridge
The genital ridge (or gonadal ridge) is the precursor to the gonads. The genital ridge initially consists mainly of mesenchyme and cells of underlying mesonephric origin. Once oogonia enter this area they attempt to associate with these somatic ce ...
. As reviewed by Messerschmidt et al., the majority of PGCs are arrested in the G2 phase of the cell cycle, while they migrate toward the hindgut during embryo days 7.5 to 8.5. Then demethylation of the PGCs takes place in two waves. At day 9.5 the primordial germ cells begin to rapidly replicate going from about 200 PGCs at embryo day 9.5 to about 10,000 PGCs at day 12.5. During days 9.5 to 12.5 DNMT3a and DNMT3b are repressed and DNMT1 is present in the nucleus at a high level. But DNMT1 is unable to methylate cytosines during days 9.5 to 12.5 because the '' UHRF1'' gene (also known as ''NP95'') is repressed and UHRF1 is an essential protein needed to recruit DNMT1 to replication foci where maintenance DNA methylation takes place. This is a passive, dilution form of demethylation.
In addition, from embryo day 9.5 to 13.5 there is an active form of demethylation. As indicated below in "Molecular stages of active reprogramming," two enzymes are central to active demethylation. These are a ten-eleven translocation methylcytosine dioxygenase (TET) and thymine-DNA glycosylase
G/T mismatch-specific thymine DNA glycosylase is an enzyme that in humans is encoded by the TDG gene. Several bacterial proteins have strong sequence homology with this protein.
Function
The protein encoded by this gene belongs to the TDG/mug ...
(TDG). One particular TET enzyme, TET1, and TDG are present at high levels from embryo day 9.5 to 13.5, and are employed in active demethylation during gametogenesis. PGC genomes display the lowest levels of DNA methylation of any cells in the entire life cycle of the mouse at embryonic day 13.5.
Learning and Memory
 Learning and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature. Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time. Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory. At 24 hours after training, 9.17% of the genes in the genomes of rat hippocampus neurons were found to be differentially methylated. This included more than 2,000 differentially methylated genes at 24 hours after training, with over 500 genes being demethylated. Similar results to that in the rat hippocampus were also obtained in mice with contextual fear conditioning.
The hippocampus region of the brain is where contextual fear memories are first stored (see figure of the brain, this section), but this storage is transient and does not remain in the hippocampus. In rats contextual fear conditioning is abolished when the hippocampus is subjected to hippocampectomy just one day after conditioning, but rats retain a considerable amount of contextual fear when hippocampectomy is delayed by four weeks. In mice, examined at 4 weeks after conditioning, the hippocampus methylations and demethylations were reversed (the hippocampus is needed to form memories but memories are not stored there) while substantial differential CpG methylation and demethylation occurred in cortical neurons during memory maintenance. There were 1,223 differentially methylated genes in the anterior cingulate cortex of mice four weeks after contextual fear conditioning. Thus, while there were many methylations in the hippocampus shortly after memory was formed, all these hippocampus methylations were demethylated as soon as four weeks later.
Learning and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature. Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time. Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory. At 24 hours after training, 9.17% of the genes in the genomes of rat hippocampus neurons were found to be differentially methylated. This included more than 2,000 differentially methylated genes at 24 hours after training, with over 500 genes being demethylated. Similar results to that in the rat hippocampus were also obtained in mice with contextual fear conditioning.
The hippocampus region of the brain is where contextual fear memories are first stored (see figure of the brain, this section), but this storage is transient and does not remain in the hippocampus. In rats contextual fear conditioning is abolished when the hippocampus is subjected to hippocampectomy just one day after conditioning, but rats retain a considerable amount of contextual fear when hippocampectomy is delayed by four weeks. In mice, examined at 4 weeks after conditioning, the hippocampus methylations and demethylations were reversed (the hippocampus is needed to form memories but memories are not stored there) while substantial differential CpG methylation and demethylation occurred in cortical neurons during memory maintenance. There were 1,223 differentially methylated genes in the anterior cingulate cortex of mice four weeks after contextual fear conditioning. Thus, while there were many methylations in the hippocampus shortly after memory was formed, all these hippocampus methylations were demethylated as soon as four weeks later.
Demethylation in Cancer
The human genome contains about 28 million CpG sites, and roughly 60% of the CpG sites are methylated at the 5 position of the cytosine. During formation of a cancer there is an average reduction of the number of methylated cytosines of about 5% to 20%, or about 840,00 to 3.4 million demethylations of CpG sites.DNMT1
DNA (cytosine-5)-methyltransferase 1 is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. In humans, it is encoded by the ''DNMT1'' gene. DNMT1 forms part of the family of ...
methylates CpGs on hemi-methylated DNA during DNA replication. Thus, when a DNA strand has a methylated CpG, and the newly replicated strand during semi-conservative replication lacks a methyl group on the complementary CpG, DNMT1 is normally recruited to the hemimethylated site and adds a methyl group to cytosine in the newly synthesized CpG. However, recruitment of DNMT1 to hemimethylated CpG sites during DNA replication depends on the UHRF1 protein. If UHRF1 does not bind to a hemimethylated CpG site, then DNMT1 is not recruited and cannot methylate the newly synthesized CpG site. The arginine methyltransferase PRMT6 regulates DNA methylation by methylating the arginine at position 2 of histone 3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a stri ...
(H3R2me2a). (See Protein methylation#Arginine.) In the presence of H3R2me2a UHRF1 can not bind to a hemimethylated CpG site, and then DNMT1 is not recruited to the site, and the site remains hemimethylated. Upon further rounds of replication the methylated CpG is passively diluted out. PRMT6 is frequently overexpressed in many types of cancer cells. The overexpression of PRMT6 may be a source of DNA demethylation in cancer.
Molecular stages of active reprogramming
Three molecular stages are required for actively, enzymatically reprogramming the DNA methylome. Stage 1: Recruitment. The enzymes needed for reprogramming are recruited to genome sites that require demethylation or methylation. Stage 2: Implementation. The initial enzymatic reactions take place. In the case of methylation, this is a short step that results in the methylation of cytosine to 5-methylcytosine. Stage 3: Base excision DNA repair. The intermediate products of demethylation are catalysed by specific enzymes of the base excision DNA repair pathway that finally restore cystosine in the DNA sequence.Stage 2 of active demethylation
TET enzymes
The TET enzymes are a family of ten-eleven translocation (TET) methylcytosine dioxygenases. They are instrumental in DNA demethylation. 5-Methylcytosine (see first Figure) is a methylated form of the DNA base cytosine (C) that often regulates ge ...
). The molecular steps of this initial demethylation are shown in detail in TET enzymes
The TET enzymes are a family of ten-eleven translocation (TET) methylcytosine dioxygenases. They are instrumental in DNA demethylation. 5-Methylcytosine (see first Figure) is a methylated form of the DNA base cytosine (C) that often regulates ge ...
. In successive steps (see Figure) TET enzymes
The TET enzymes are a family of ten-eleven translocation (TET) methylcytosine dioxygenases. They are instrumental in DNA demethylation. 5-Methylcytosine (see first Figure) is a methylated form of the DNA base cytosine (C) that often regulates ge ...
further hydroxylate 5hmC to generate 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Thymine-DNA glycosylase
G/T mismatch-specific thymine DNA glycosylase is an enzyme that in humans is encoded by the TDG gene. Several bacterial proteins have strong sequence homology with this protein.
Function
The protein encoded by this gene belongs to the TDG/mug ...
(TDG) recognizes the intermediate bases 5fC and 5caC and excises the glycosidic bond resulting in an apyrimidinic site (AP site). This is followed by base excision repair (stage 3). In an alternative oxidative deamination pathway, 5hmC can be oxidatively deaminated by APOBEC
image:Apobec.J.Steinfeld.D.png, 300px, upExample of a member of the APOBEC family, APOBEC-2. A cytidine deaminase from ''Homo sapiens''.; ; rendered usinPyMOL
APOBEC ("apolipoprotein B mRNA editing enzyme, catalytic polypeptide") is a family o ...
(AID/APOBEC) deaminases to form 5-hydroxymethyluracil (5hmU). Also, 5mC can be converted to thymine
Thymine () ( symbol T or Thy) is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidi ...
(Thy). 5hmU can be cleaved by TDG, MBD4, NEIL1 or SMUG1
Single-strand selective monofunctional uracil DNA glycosylase is an enzyme that in humans is encoded by the ''SMUG1'' gene. SMUG1 is a glycosylase that removes uracil from single- and double-stranded DNA in nuclear chromatin, thus contributing to ...
. AP sites and T:G mismatches are then repaired by base excision repair (BER) enzymes to yield cytosine (Cyt). The TET family of dioxygenases are employed in the most frequent type of demethylation reactions.
TET family
TET dioxygenase isoforms include at least two isoforms of TET1, one of TET2 and three isoforms of TET3. The full-length canonical TET1 isoform appears virtually restricted to early embryos, embryonic stem cells and primordial germ cells (PGCs). The dominant TET1 isoform in most somatic tissues, at least in the mouse, arises from alternative promoter usage which gives rise to a short transcript and a truncated protein designated TET1s. The isoforms of TET3 are the full length form TET3FL, a short form splice variant TET3s, and a form that occurs in oocytes and neurons designated TET3o. TET3o is created by alternative promoter use and contains an additional first N-terminal exon coding for 11 amino acids. TET3o only occurs in oocytes and neurons and is not expressed in embryonic stem cells or in any other cell type or adult mouse tissue tested. Whereas TET1 expression can barely be detected in oocytes and zygotes, and TET2 is only moderately expressed, the TET3 variant TET3o shows extremely high levels of expression in oocytes and zygotes, but is nearly absent at the 2-cell stage. It is possible that TET3o, high in neurons, oocytes and zygotes at the one cell stage, is the major TET enzyme utilized when very large scale rapid demethylations occur in these cells.Stage 1 of demethylation - recruitment of TET to DNA
The TET enzymes do not specifically bind to 5-methylcytosine except when recruited. Without recruitment or targeting, TET1 predominantly binds to high CG promoters and CpG islands (CGIs) genome-wide by its CXXC domain that can recognize un-methylated CGIs. TET2 does not have an affinity for 5-methylcytosine in DNA. The CXXC domain of the full-length TET3, which is the predominant form expressed in neurons, binds most strongly to CpGs where the C was converted to 5-carboxycytosine (5caC). However, it also binds to un-methylated CpGs.CpG site
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG isl ...
in DNA. Two of the proteins shown to recruit a TET enzyme to a methylated cytosine in DNA are OGG1 (see figure Initiation of DNA demethylation at a CpG site) and EGR1
EGR-1 (Early growth response protein 1) also known as ZNF268 (zinc finger protein 268) or NGFI-A (nerve growth factor-induced protein A) is a protein that in humans is encoded by the ''EGR1'' gene.
EGR-1 is a mammalian transcription factor. It wa ...
.
OGG1
Oxoguanine glycosylase (OGG1) catalyses the first step in base excision repair of the oxidatively damaged base 8-OHdG. OGG1 finds 8-OHdG by sliding along the linear DNA at 1,000 base pairs of DNA in 0.1 seconds. OGG1 very rapidly finds 8-OHdG. OGG1 proteins bind to oxidatively damaged DNA with a half maximum time of about 6 seconds. When OGG1 finds 8-OHdG it changes conformation and complexes with 8-OHdG in its binding pocket. OGG1 does not immediately act to remove the 8-OHdG. Half maximum removal of 8-OHdG takes about 30 minutes in HeLa cells ''in vitro'', or about 11 minutes in the livers of irradiated mice. DNA oxidation by reactive oxygen species preferentially occurs at a guanine in a methylated CpG site, because of a lowered ionization potential of guanine bases adjacent to 5-methylcytosine. TET1 binds (is recruited to) the OGG1 bound to 8-OHdG (see figure). This likely allows TET1 to demethylate an adjacent methylated cytosine. When human mammary epithelial cells (MCF-10A) were treated with H2O2, 8-OHdG increased in DNA by 3.5-fold and this caused about 80% demethylation of the 5-methylcytosines in the MCF-10A genome.EGR1
The gene ''early growth response protein 1'' (''EGR1
EGR-1 (Early growth response protein 1) also known as ZNF268 (zinc finger protein 268) or NGFI-A (nerve growth factor-induced protein A) is a protein that in humans is encoded by the ''EGR1'' gene.
EGR-1 is a mammalian transcription factor. It wa ...
'') is an immediate early gene
Immediate early genes (IEGs) are genes which are activated transiently and rapidly in response to a wide variety of cellular stimuli. They represent a standing response mechanism that is activated at the transcription level in the first round of ...
(IEG). EGR1 can rapidly be induced by neuronal activity. The defining characteristic of IEGs is the rapid and transient up-regulation—within minutes—of their mRNA levels independent of protein synthesis. In adulthood, EGR1 is expressed widely throughout the brain, maintaining baseline expression levels in several key areas of the brain including the medial prefrontal cortex, striatum, hippocampus and amygdala. This expression is linked to control of cognition, emotional response, social behavior and sensitivity to reward. EGR1 binds to DNA at sites with the motifs 5′-GCGTGGGCG-3′ and 5'-GCGGGGGCGG-3′ and these motifs occur primarily in promoter regions of genes. The short isoform TET1s is expressed in the brain. EGR1 and TET1s form a complex mediated by the C-terminal regions of both proteins, independently of association with DNA. EGR1 recruits TET1s to genomic regions flanking EGR1 binding sites. In the presence of EGR1, TET1s is capable of locus-specific demethylation and activation of the expression of downstream genes regulated by EGR1.
DNA demethylation intermediate 5hmC
As indicated in the Figure above, captioned "Demethylation of 5-methylcytosine," the first step in active demethylation is a TET oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). The demethylation process, in some tissues and at some genome locations, may stop at that point. As reviewed by Uribe-Lewis et al., in addition to being an intermediate in active DNA demethylation, 5hmC is often a stable DNA modification. Within the genome, 5hmC is located at transcriptionally active genes, regulatory elements and chromatin associated complexes. In particular, 5hmC is dynamically changed and positively correlated with active gene transcription duringcell lineage
Cell lineage denotes the developmental history of a tissue or organ from the fertilized embryo. This is based on the tracking of an organism's cellular ancestry due to the cell divisions and relocation as time progresses, this starts with the orig ...
specification, and high levels of 5hmC are found in embryonic stem cell
Embryonic stem cells (ESCs) are pluripotent stem cells derived from the inner cell mass of a blastocyst, an early-stage pre- implantation embryo. Human embryos reach the blastocyst stage 4–5 days post fertilization, at which time they consist ...
s and in the central nervous system
The central nervous system (CNS) is the part of the nervous system consisting primarily of the brain and spinal cord. The CNS is so named because the brain integrates the received information and coordinates and influences the activity of all p ...
. In humans, defective 5-hydroxymethylating activity is associated with a phenotype of lymphoproliferation, immunodeficiency and autoimmunity.
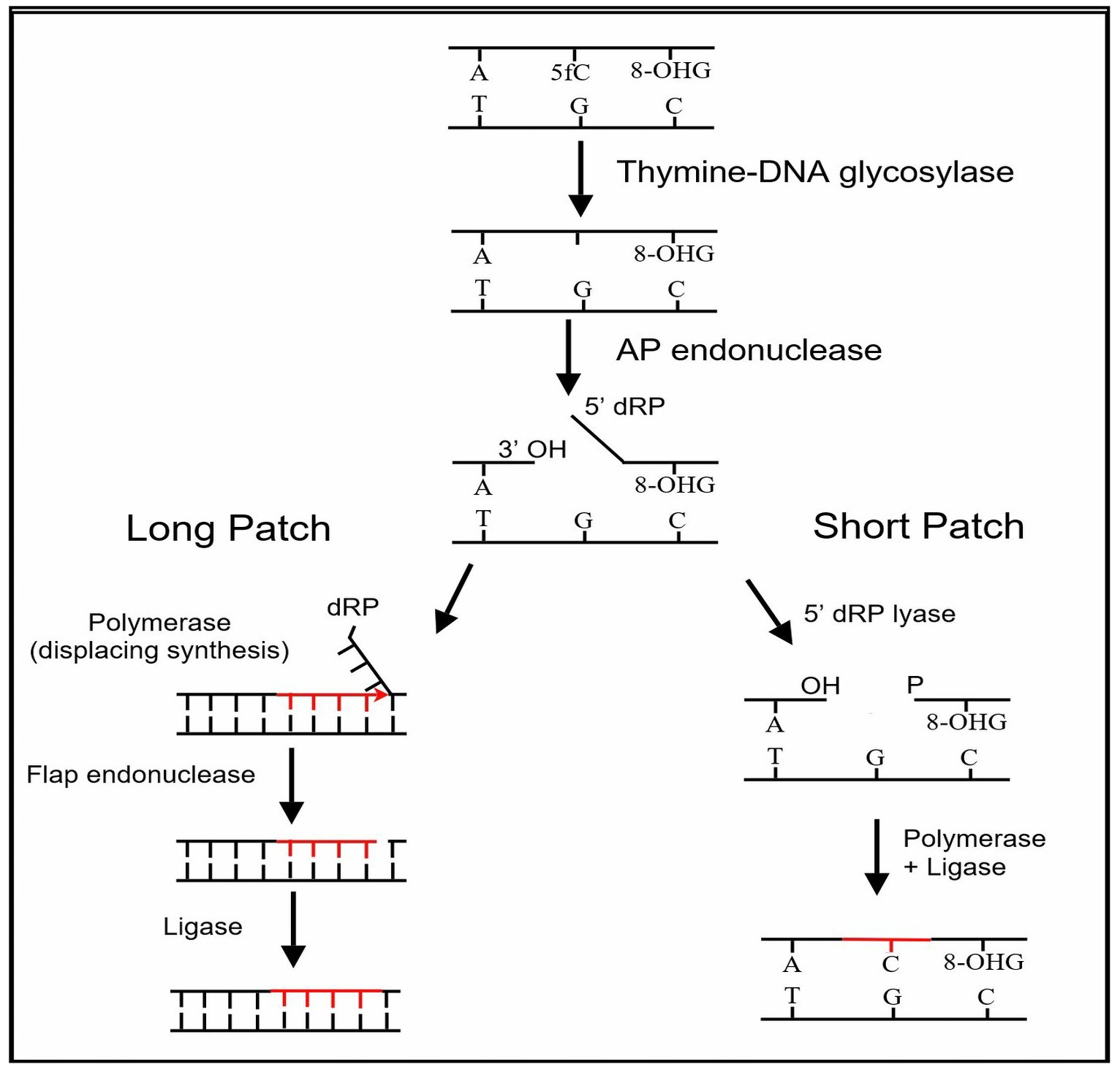
Stage 3 base excision repair
 The third stage of DNA demethylation is removal of the intermediate products of demethylation generated by a TET enzyme by
The third stage of DNA demethylation is removal of the intermediate products of demethylation generated by a TET enzyme by base excision repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from t ...
. As indicated above in Stage 2, after 5mC is first oxidized by a TET to form 5hmC, further oxidation of 5hmC by TET yields 5fC and oxidation of 5fC by TET yields 5caC. Both 5fC and 5caC are recognized by a DNA glycosylase
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first st ...
, TDG, a base excision repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from t ...
enzyme, as an abnormal base. As shown in the Figure in this section, TDG removes the abnormal base (e.g. 5fC) while leaving the sugar-phosphate backbone intact, creating an apurinic/apyrimidinic site, commonly referred to as an AP site
In biochemistry and molecular genetics, an AP site (apurinic/apyrimidinic site), also known as an abasic site, is a location in DNA (also in RNA but much less likely) that has neither a purine nor a pyrimidine base, either spontaneously or due ...
. In this Figure, the 8-OHdG is left in the DNA, since it may have been present when OGG1 attracted TET1 to the CpG site with a methylated cytosine. After an AP site is formed, AP endonuclease
Apurinic/apyrimidinic (AP) endonuclease is an enzyme that is involved in the DNA base excision repair pathway (BER). Its main role in the repair of damaged or mismatched nucleotides in DNA is to create a nick in the phosphodiester backbone of t ...
creates a nick in the phosphodiester
In chemistry, a phosphodiester bond occurs when exactly two of the hydroxyl groups () in phosphoric acid react with hydroxyl groups on other molecules to form two ester bonds. The "bond" involves this linkage . Discussion of phosphodiesters is d ...
backbone of the AP site
In biochemistry and molecular genetics, an AP site (apurinic/apyrimidinic site), also known as an abasic site, is a location in DNA (also in RNA but much less likely) that has neither a purine nor a pyrimidine base, either spontaneously or due ...
that was formed when the TDG DNA glycosylase
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first st ...
removed the 5fC or 5caC. The human AP endonuclease incises DNA 5′ to the AP site by a hydrolytic mechanism, leaving a 3′-hydroxyl and a 5′-deoxyribose phosphate (5' dRP) residue. This is followed by either short patch or long patch repair. In short patch repair, 5′ dRP lyase trims the 5′ dRP end to form a phosphorylated 5′ end. This is followed by DNA polymerase β (pol β) adding a single cytosine to pair with the pre-existing guanine in the complementary strand and then DNA ligase
DNA ligase is a specific type of enzyme, a ligase, () that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living orga ...
to seal the cut strand. In long patch repair, DNA synthesis is thought to be mediated by polymerase δ and polymerase ε performing displacement synthesis to form a flap. Pol β can also perform long-patch displacement synthesis. Long-patch synthesis typically inserts 2–10 new nucleotides. Then flap endonuclease removes the flap, and this is followed by DNA ligase
DNA ligase is a specific type of enzyme, a ligase, () that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living orga ...
to seal the strand. At this point there has been a complete replacement of the 5-methylcytosine by cytosine (demethylation) in the DNA sequence.
Demethylation after exercise
Physical exercise has well established beneficial effects on learning and memory (seeNeurobiological effects of physical exercise
The neurobiological effects of physical exercise are numerous and involve a wide range of interrelated effects on brain structure, brain function, and cognition. A large body of research in humans has demonstrated that consistent aerobic exer ...
). ''BDNF
Brain-derived neurotrophic factor (BDNF), or abrineurin, is a protein found in the and the periphery. that, in humans, is encoded by the ''BDNF'' gene. BDNF is a member of the neurotrophin family of growth factors, which are related to the canon ...
'' is a particularly important regulator of learning and memory. As reviewed by Fernandes et al., in rats, exercise enhances the hippocampus
The hippocampus (via Latin from Greek , ' seahorse') is a major component of the brain of humans and other vertebrates. Humans and other mammals have two hippocampi, one in each side of the brain. The hippocampus is part of the limbic system, ...
expression of the gene ''Bdnf
Brain-derived neurotrophic factor (BDNF), or abrineurin, is a protein found in the and the periphery. that, in humans, is encoded by the ''BDNF'' gene. BDNF is a member of the neurotrophin family of growth factors, which are related to the canon ...
'', which has an essential role in memory formation. Enhanced expression of ''Bdnf'' occurs through demethylation of its CpG island promoter at exon IV and this demethylation depends on steps illustrated in the two figures.
Demethylation after exposure to traffic related air pollution
In a panel of healthy adults, negative associations were found between total DNA methylation and exposure to traffic related air pollution. DNA methylation levels were associated both with recent and chronic exposure to Black Carbon as well as benzene.Peripheral sensory neuron regeneration
After injury,neuron
A neuron, neurone, or nerve cell is an electrically excitable cell that communicates with other cells via specialized connections called synapses. The neuron is the main component of nervous tissue in all animals except sponges and placozoa. ...
s in the adult peripheral nervous system
The peripheral nervous system (PNS) is one of two components that make up the nervous system of bilateral animals, with the other part being the central nervous system (CNS). The PNS consists of nerves and ganglia, which lie outside the brain ...
can switch from a dormant state with little axon
An axon (from Greek ἄξων ''áxōn'', axis), or nerve fiber (or nerve fibre: see spelling differences), is a long, slender projection of a nerve cell, or neuron, in vertebrates, that typically conducts electrical impulses known as action p ...
al growth to robust axon regeneration. DNA demethylation in mature mammalian neurons removes barriers to axonal regeneration. This demethylation, in regenerating mouse peripheral neurons, depends upon TET3 to generate 5-hydroxymethylcytosine (5hmC) in DNA. 5hmC was altered in a large set of regeneration-associated genes (RAGs), including well-known RAGs such as '' Atf3'', ''Bdnf
Brain-derived neurotrophic factor (BDNF), or abrineurin, is a protein found in the and the periphery. that, in humans, is encoded by the ''BDNF'' gene. BDNF is a member of the neurotrophin family of growth factors, which are related to the canon ...
'', and ''Smad1
Mothers against decapentaplegic homolog 1 also known as SMAD family member 1 or SMAD1 is a protein that in humans is encoded by the ''SMAD1'' gene.
Nomenclature
SMAD1 belongs to the SMAD, a family of proteins similar to the gene products of the ...
'', that regulate the axon growth potential of neurons.
References
{{DEFAULTSORT:Dna Demethylation Molecular biology