|
De Novo Gene Birth
''De novo'' gene birth is the process by which new genes evolve from non-coding DNA. '' De novo'' genes represent a subset of novel genes, and may be protein-coding or instead act as RNA genes. The processes that govern ''de novo'' gene birth are not well understood, although several models exist that describe possible mechanisms by which ''de novo'' gene birth may occur. Although ''de novo'' gene birth may have occurred at any point in an organism's evolutionary history, ancient ''de novo'' gene birth events are difficult to detect. Most studies of ''de novo'' genes to date have thus focused on young genes, typically taxonomically restricted genes (TRGs) that are present in a single species or lineage, including so-called orphan genes, defined as genes that lack any identifiable homolog. It is important to note, however, that not all orphan genes arise ''de novo'', and instead may emerge through fairly well characterized mechanisms such as gene duplication (including retropo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriophage
A bacteriophage (), also known informally as a phage (), is a virus that infects and replicates within bacteria. The term is derived . Bacteriophages are composed of proteins that Capsid, encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes (e.g. Bacteriophage MS2, MS2) and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm. Bacteriophages are among the most common and diverse entities in the biosphere. Bacteriophages are ubiquitous viruses, found wherever bacteria exist. It is estimated there are more than 1031 bacteriophages on the planet, more than every other organism on Earth, including bacteria, combined. Viruses are the most abundant biological entity in the water column of the world's oceans, and the second largest component of biomass after prokaryotes, where up to 9x108 virus, virions per millilitre have b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
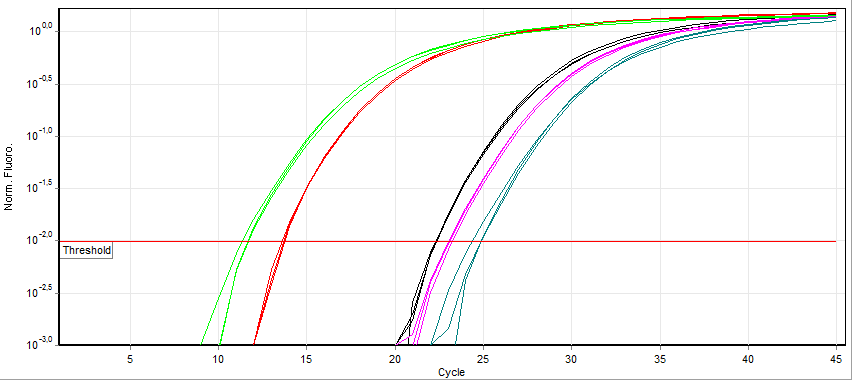
Quantitative PCR
A real-time polymerase chain reaction (real-time PCR, or qPCR when used quantitatively) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively (i.e., above/below a certain amount of DNA molecules). Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter, which permits detection only after hybridization of the probe with its complementary sequence. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines propose that the abbreviation ''qPCR'' be used for quantitative real-time PCR and that ' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BLAST (biotechnology)
In bioinformatics, BLAST (basic local alignment search tool) is an algorithm and program for comparing Primary structure, primary biological sequence information, such as the amino acid, amino-acid sequences of proteins or the nucleotides of DNA sequence, DNA and/or RNA sequences. A BLAST search enables a researcher to compare a subject protein or nucleotide sequence (called a query) with a library or database of sequences, and identify database sequences that resemble the query sequence above a certain threshold. For example, following the discovery of a previously unknown gene in the Mus musculus, mouse, a scientist will typically perform a BLAST search of the human genome to see if humans carry a similar gene; BLAST will identify sequences in the human genome that resemble the mouse gene based on similarity of sequence. Background BLAST is one of the most widely used bioinformatics programs for sequence searching. It addresses a fundamental problem in bioinformatics research ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Homology
Sequence homology is the homology (biology), biological homology between DNA sequence, DNA, RNA sequence, RNA, or Protein primary structure, protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a Gene duplication, duplication event (paralogs), or else a Horizontal gene transfer, horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Sequence alignment, Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Synteny
In genetics, the term synteny refers to two related concepts: * In classical genetics, ''synteny'' describes the physical co-localization of genetic loci on the same chromosome within an individual or species. * In current biology, ''synteny'' more commonly refers to ''colinearity'', i.e. conservation of blocks of order within two sets of chromosomes that are being compared with each other. These blocks are referred to as ''syntenic blocks''. The Encyclopædia Britannica gives the following description of synteny, using the modern definition: Etymology ''Synteny'' is a neologism meaning "on the same ribbon"; Greek: ', ''syn'' "along with" + ', ''tainiā'' "band". This can be interpreted classically as "on the same chromosome", or in the modern sense of having the same order of genes on two (homologous) strings of DNA (or chromosomes). : co-localization on a chromosome The classical concept is related to genetic linkage: Linkage between two loci is established by the observat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
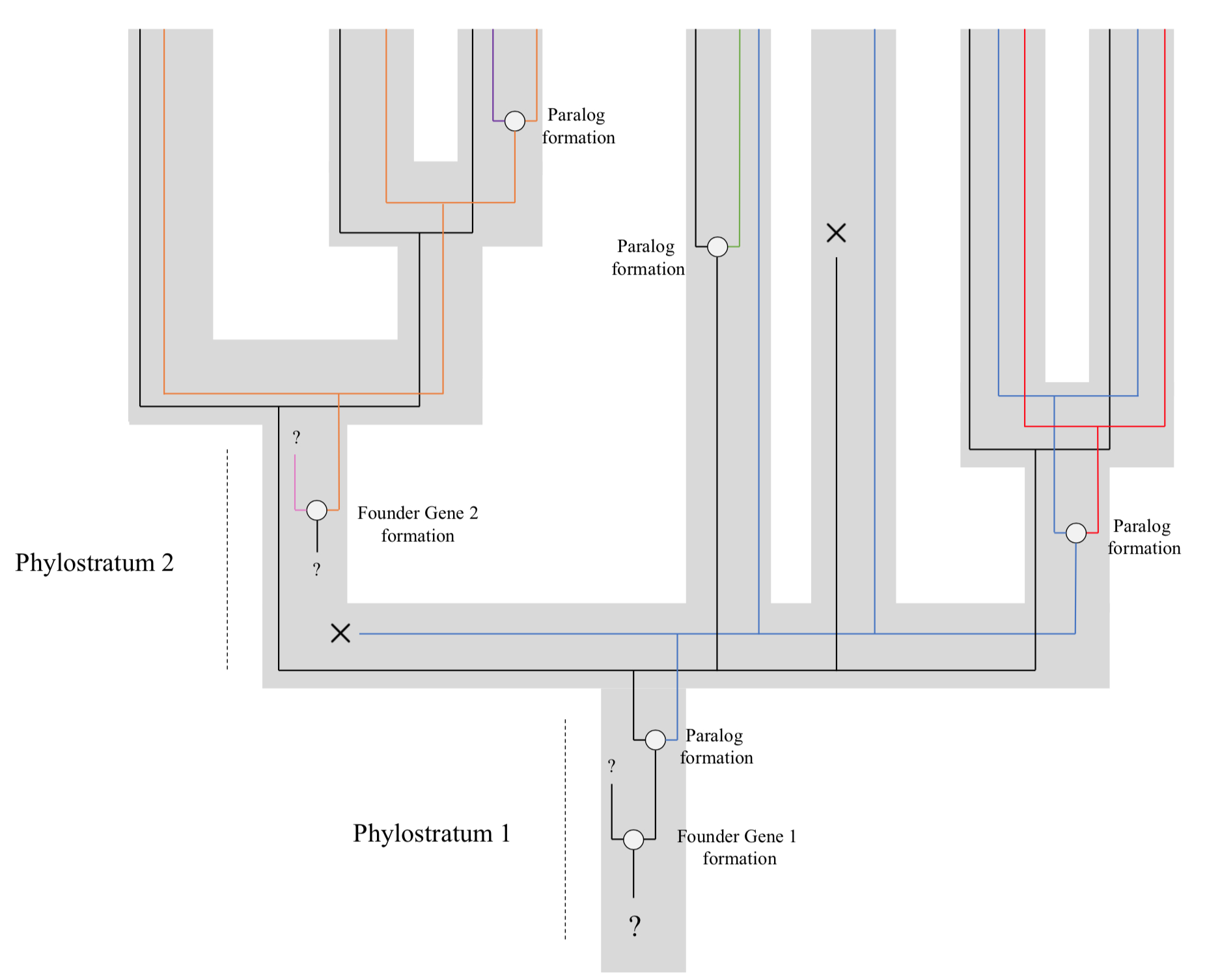
Genomic Phylostratigraphy
Genomic phylostratigraphy is a novel genetic statistical method developed in order to date the origin of specific genes by looking at its homologs across species. It was first developed by Ruđer Bošković Institute in Zagreb, Croatia. The system links genes to their founder gene, allowing us to then determine their age. This could help us better understand many evolutionary processes such as patterns of gene birth throughout evolution, or the relationship between the age of a transcriptome throughout embryonic development. Bioinformatic tools likGenErahave been developed to calculate relative gene ages based on genomic phylostratigraphy. Method This technique relies on the assumption that the diversity of the genome is not only due to gene duplications but also to continuous frequent de novo gene births. These genes (called "founder genes") would form from non-genic DNA sequences, as well as from changes in reading frame (or other ways of arising from within existing genes), ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Orphan Gene
Orphan genes, ORFans, or taxonomically restricted genes (TRGs) are genes that lack a detectable homologue outside of a given species or lineage. Most genes have known homologues. Two genes are homologous when they share an evolutionary history, and the study of groups of homologous genes allows for an understanding of their evolutionary history and divergence. Common mechanisms that have been uncovered as sources for new genes through studies of homologues include gene duplication, exon shuffling, gene fusion and fission, etc. Studying the origins of a gene becomes more difficult when there is no evident homologue. The discovery that about 10% or more of the genes of the average microbial species is constituted by orphan genes raises questions about the evolutionary origins of different species as well as how to study and uncover the evolutionary origins of orphan genes. In some cases, a gene can be classified as an orphan gene due to undersampling of the existing genome space. W ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Arabidopsis Thaliana
''Arabidopsis thaliana'', the thale cress, mouse-ear cress or arabidopsis, is a small plant from the mustard family (Brassicaceae), native to Eurasia and Africa. Commonly found along the shoulders of roads and in disturbed land, it is generally considered a weed. A winter annual with a relatively short lifecycle, ''A. thaliana'' is a popular model organism in plant biology and genetics. For a complex multicellular eukaryote, ''A. thaliana'' has a relatively small genome of around 135 Base pair#Length measurements, megabase pairs. It was the first plant to have its genome sequenced, and is an important tool for understanding the molecular biology of many plant traits, including flower development and phototropism, light sensing. Description ''Arabidopsis thaliana'' is an annual plant, annual (rarely biennial plant, biennial) plant, usually growing to 20–25 cm tall. The leaf, leaves form a rosette at the base of the plant, with a few leaves also on the flowering Plant ste ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosophila Simulans
''Drosophila simulans'' is a species of fly closely related to '' D. melanogaster'', belonging to the same ''melanogaster'' species subgroup. Its closest relatives are ''D. mauritiana'' and ''D. sechellia''. Taxonomy This species was discovered by the fly geneticist Alfred Sturtevant in 1919, when he noticed that the flies used in Thomas Hunt Morgan's laboratory at the Columbia University were actually two distinct species: '' D. melanogaster'' and ''D. simulans''. Males differ in the external genitalia, while trained observers can separate females using colour characteristics. ''D. melanogaster'' females crossed to ''D. simulans'' males produce sterile F1 females and no F1 males. The reciprocal cross produces sterile F1 males and no female progeny. ''Drosophila simulans'' was found later to be closely related to two island endemics, ''D. sechellia'' and ''D. mauritiana''. ''D. simulans'' will mate with these sister species to form fertile females and sterile males, a fact t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosophila Melanogaster
''Drosophila melanogaster'' is a species of fly (an insect of the Order (biology), order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the "vinegar fly", "pomace fly", or "banana fly". In the wild, ''D. melanogaster'' are attracted to rotting fruit and fermenting beverages, and are often found in orchards, kitchens and pubs. Starting with Charles W. Woodworth's 1901 proposal of the use of this species as a model organism, ''D. melanogaster'' continues to be widely used for biological research in genetics, physiology, microbial pathogenesis, and Life history theory, life history evolution. ''D. melanogaster'' was the first animal to be Fruit flies in space, launched into space in 1947. As of 2017, six Nobel Prizes have been awarded to drosophilists for their work using the insect. ''Drosophila melanogaster'' is typically used in research owing to its rapid life cycle, relatively simple genetics with on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosophila Erecta
''Drosophila erecta'' is a West African species A species () is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. It is the basic unit of Taxonomy (biology), ... of fruit fly, and was one of 12 fruit fly genomes sequenced for a large comparative study. References External links ''Drosophila erecta'' at FlyBase''Drosophila erecta'' at Ensembl Genomes Metazoa* e {{Drosophilidae-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |