|
TEC-RED
Trans-Spliced Exon Coupled RNA End Determination (TEC-RED) is a transcriptomic technique that, like SAGE, allows for the digital detection of messenger RNA sequences. Unlike SAGE, detection and purification of transcripts from the 5’ end of the messenger RNA require the presence of a trans-spliced leader sequence. Trans-splicing Background Spliced leader sequences are short sequences of non coding RNA, not found within a gene itself, that are attached to the 5’ end of all, or a portion of, mRNAs transcribed in an organism. They have been found in several species to be responsible for separating polycistronic transcripts into single gene mRNAs, and in others to splice onto monocistronic transcripts. The major role of trans-splicing on monocistronic transcripts is largely unknown. It has been proposed that they may act as an independent promoter that aids in tissue specific expression of independent protein isoforms. Spliced leaders have been seen in trypanosomatids, Euglena, f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcriptomics Technologies
Transcriptomics technologies are the techniques used to study an organism's transcriptome, the sum of all of its RNA transcripts. The information content of an organism is recorded in the DNA of its genome and expressed through transcription. Here, mRNA serves as a transient intermediary molecule in the information network, whilst non-coding RNAs perform additional diverse functions. A transcriptome captures a snapshot in time of the total transcripts present in a cell. Transcriptomics technologies provide a broad account of which cellular processes are active and which are dormant. A major challenge in molecular biology is to understand how a single genome gives rise to a variety of cells. Another is how gene expression is regulated. The first attempts to study whole transcriptomes began in the early 1990s. Subsequent technological advances since the late 1990s have repeatedly transformed the field and made transcriptomics a widespread discipline in biological sciences. There a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research and forensic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also known as transcriptome. RNA-Seq facilitates the ability to look at alternative gene spliced transcripts, post-transcriptional modifications, gene fusion, mutations/ SNPs and changes in gene expression over time, or differences in gene expression in different groups or treatments. In addition to mRNA transcripts, RNA-Seq can look at different populations of RNA to include total RNA, small RNA, such as miRNA, tRNA, and ribosomal profiling. RNA-Seq can also be used to determine exon/intron boundaries and verify or amend previously annotated 5' and 3' gene boundaries. Recent advances in RNA-Seq include single cell sequencing, bulk RNA sequencing, 3' mRNA-sequencing, in situ sequencing of fixed tissue, and native RNA molecule sequencin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
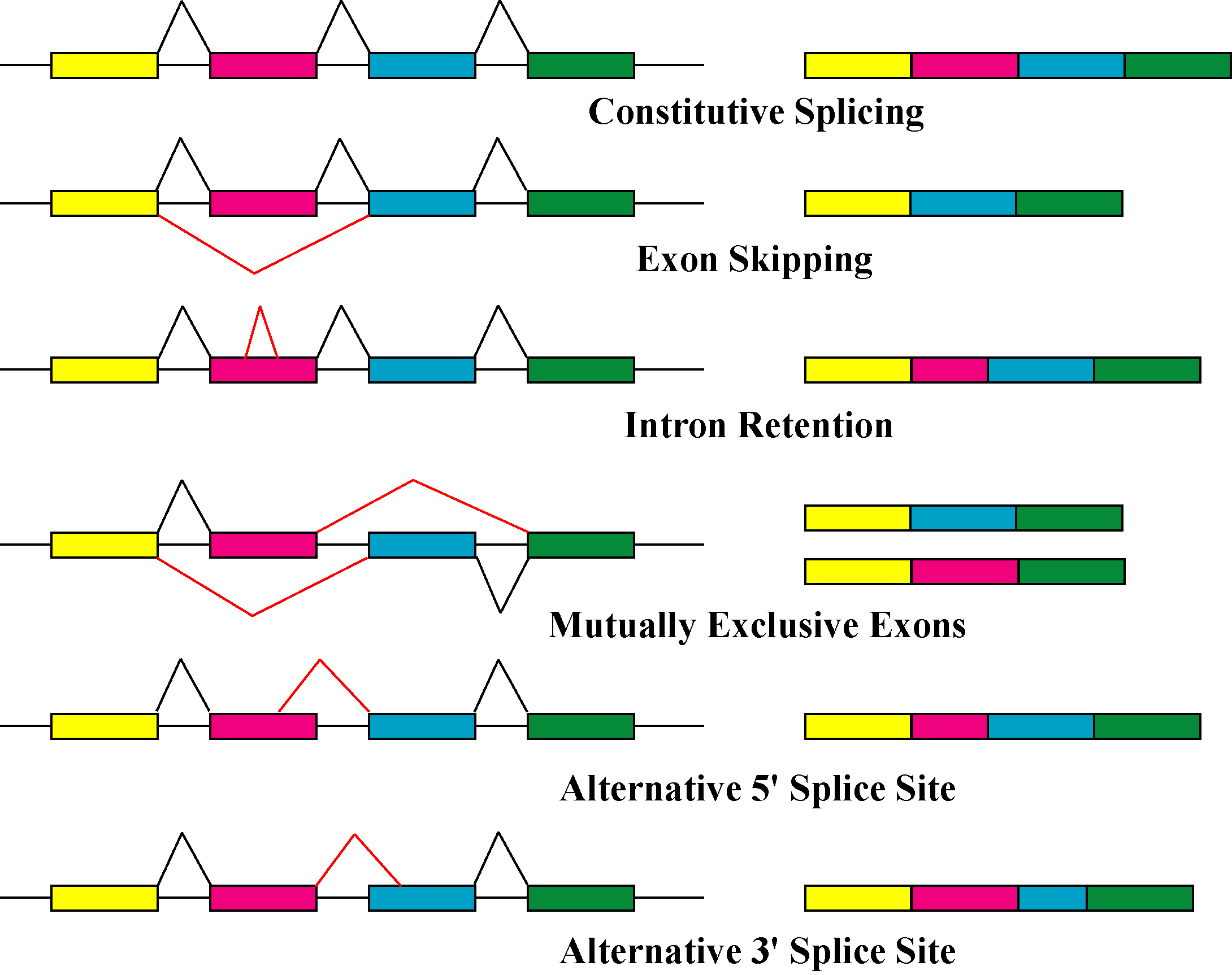
Alternative Splicing
Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative RNA splicing, splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene may be included within or excluded from the final RNA product of the gene. This means the exons are joined in different combinations, leading to different splice variants. In the case of protein-coding genes, the proteins translated from these splice variants may contain differences in their amino acid sequence and in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most of the observed splice variants ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isoforms
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions of genes revealed by the human genome project and the large diversity of proteins seen in an organism: different proteins enc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TEC RED ConcatenationB
TEC or Tec may refer to: Education * Trichy Engineering College, in Konalai, Tamil Nadu, India * Costa Rica Institute of Technology * Tec, the Monterrey Institute of Technology and Higher Education * Tertiary Education Commission (other) * Texas Eastern Conference, an American junior college athletic conference Governmental and public organizations * Tasmanian Electoral Commission, Australia * The Episcopal Church * Telecommunication Engineering Centre, an Indian government agency * Tsunami Evaluation Coalition * Transatlantic Economic Council, directs economic cooperation between the US and the EU * Training and enterprise council, former public bodies in th UK * Topographic Engineering Center, former name of the US Army Geospatial Center * Treaty establishing the European Community * Transitional Executive Council, historic South African organization Military * TEC-3 or Technician third grade, United States Army rank * TEC-4 or Technician fourth grade, United S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid Vector
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and archaea; however plasmids are sometimes present in and eukaryotic organisms as well. Plasmids often carry useful genes, such as those involved in antibiotic resistance, virulence, secondary metabolism and bioremediation. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain additional genes for special circumstances. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet by various vendors using ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Ligase
DNA ligase is a type of enzyme that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms (such as DNA ligase IV) may specifically repair double-strand breaks (i.e. a break in both complementary strands of DNA). Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA. DNA ligase is used in both DNA repair and DNA replication (see '' Mammalian ligases''). In addition, DNA ligase has extensive use in molecular biology laboratories for recombinant DNA experiments (see '' Research applications''). Purified DNA ligase is used in gene cloning to join DNA molecules together to form recombinant DNA. Enzymatic mechanism The mechanism of DNA ligase is to form two covalent phosphodiester bonds between ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biotin
Biotin (also known as vitamin B7 or vitamin H) is one of the B vitamins. It is involved in a wide range of metabolic processes, both in humans and in other organisms, primarily related to the utilization of fats, carbohydrates, and amino acids. The name ''biotin'', borrowed from the German , derives from the Ancient Greek word (; 'life') and the suffix "-in" (a suffix used in chemistry usually to indicate 'forming'). Biotin appears as a white, needle-like crystalline solid. Chemical description Biotin is classified as a heterocyclic compound, with a sulfur-containing tetrahydrothiophene ring fused to a ureido group. A C5-carboxylic acid side chain is appended to the former ring. The ureido ring, containing the −N−CO−N− group, serves as the carbon dioxide carrier in carboxylation reactions. Biotin is a coenzyme for five carboxylase enzymes, which are involved in the catabolism of amino acids and fatty acids, synthesis of fatty acids, and gluconeogenesis. Biotinylat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Endonuclease
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA enzyme substrate (biology), substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each backbone chain, sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CDNA
In genetics, complementary DNA (cDNA) is DNA that was reverse transcribed (via reverse transcriptase) from an RNA (e.g., messenger RNA or microRNA). cDNA exists in both single-stranded and double-stranded forms and in both natural and engineered forms. In engineered forms, it often is a copy (replicate) of the naturally occurring DNA from any particular organism's natural genome; the organism's own mRNA was naturally transcribed from its DNA, and the cDNA is reverse transcribed from the mRNA, yielding a duplicate of the original DNA. Engineered cDNA is often used to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), or to sequence or quantify mRNA molecules using DNA based methods (qPCR, RNA-seq). cDNA that codes for a specific protein can be transferred to a recipient cell for expression as part of recombinant DNA, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serial Analysis Of Gene Expression
Serial Analysis of Gene Expression (SAGE) is a transcriptomic technique used by molecular biologists to produce a snapshot of the messenger RNA population in a sample of interest in the form of small tags that correspond to fragments of those transcripts. Several variants have been developed since, most notably a more robust version, LongSAGE, RL-SAGE and the most recent SuperSAGE. Many of these have improved the technique with the capture of longer tags, enabling more confident identification of a source gene. Overview Briefly, SAGE experiments proceed as follows: # The mRNA of an input sample (e.g. a tumour) is isolated and a reverse transcriptase and biotinylated primers are used to synthesize cDNA from mRNA. # The cDNA is bound to Streptavidin beads via interaction with the biotin attached to the primers, and is then cleaved using a restriction endonuclease called an anchoring enzyme (AE). The location of the cleavage site and thus the length of the remaining cDNA bound ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |