|
Sequence Saturation Mutagenesis
Sequence saturation mutagenesis (SeSaM) is a chemo-enzymatic random mutagenesis method applied for the directed evolution of proteins and enzymes. It is one of the most common saturation mutagenesis techniques. In four PCR-based reaction steps, phosphorothioate nucleotides are inserted in the gene sequence, cleaved and the resulting fragments elongated by universal or degenerate nucleotides. These nucleotides are then replaced by standard nucleotides, allowing for a broad distribution of nucleic acid mutations spread over the gene sequence with a preference to transversions and with a unique focus on consecutive point mutations, both difficult to generate by other mutagenesis techniques. The technique was developed by Professor Ulrich Schwaneberg at Jacobs University Bremen and RWTH Aachen University. Technology, development and advantages SeSaM has been developed in order to overcome several of the major limitations encountered when working with standard mutagenesis methods bas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mutagenesis (molecular Biology Technique)
Mutagenesis () is a process by which the genetic information of an organism is changed by the production of a mutation. It may occur spontaneously in nature, or as a result of exposure to mutagens. It can also be achieved experimentally using laboratory procedures. A mutagen is a mutation-causing agent, be it chemical or physical, which results in an increased rate of mutations in an organism's genetic code. In nature mutagenesis can lead to cancer and various heritable diseases, and it is also a driving force of evolution. Mutagenesis as a science was developed based on work done by Hermann Joseph Muller, Hermann Muller, Charlotte Auerbach and J. M. Robson in the first half of the 20th century. History DNA may be modified, either naturally or artificially, by a number of physical, chemical and biological agents, resulting in mutations. Hermann Joseph Muller, Hermann Muller found that "high temperatures" have the ability to mutate genes in the early 1920s, and in 1927, demonstrated ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
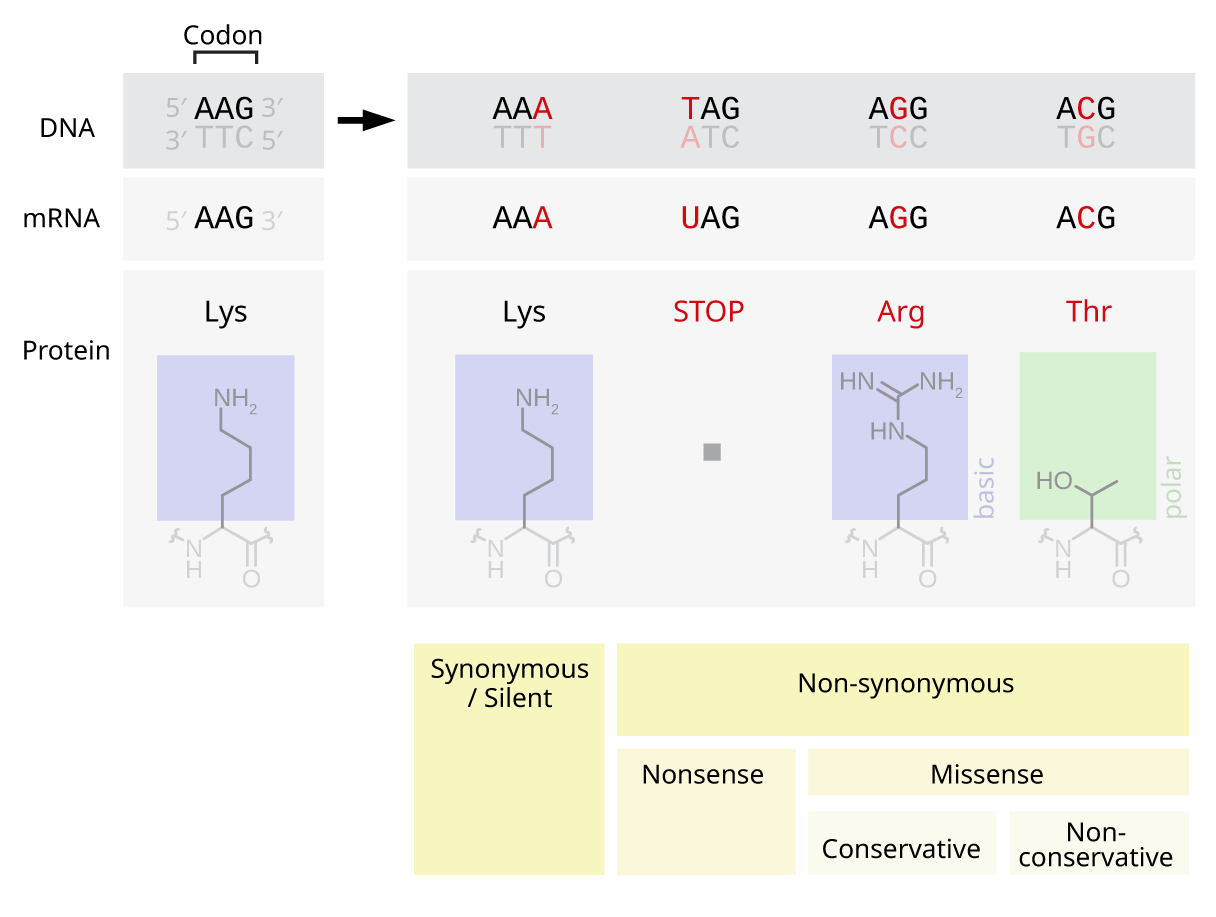
Codon Degeneracy
Degeneracy or redundancy of codons is the redundancy of the genetic code, exhibited as the multiplicity of three-base pair codon combinations that specify an amino acid. The degeneracy of the genetic code is what accounts for the existence of synonymous mutations. Background Degeneracy of the genetic code was identified by Lagerkvist. For instance, codons GAA and GAG both specify glutamic acid and exhibit redundancy; but, neither specifies any other amino acid and thus are not ambiguous or demonstrate no ambiguity. The codons encoding one amino acid may differ in any of their three positions; however, more often than not, this difference is in the second or third position. For instance, the amino acid glutamic acid is specified by GAA and GAG codons (difference in the third position); the amino acid leucine is specified by UUA, UUG, CUU, CUC, CUA, CUG codons (difference in the first or third position); and the amino acid serine is specified by UCA, UCG, UCC, UCU, AGU, AGC (differe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Genetics
Molecular genetics is a branch of biology that addresses how differences in the structures or expression of DNA molecules manifests as variation among organisms. Molecular genetics often applies an "investigative approach" to determine the structure and/or function of genes in an organism's genome using genetic screens. The field of study is based on the merging of several sub-fields in biology: classical Mendelian inheritance, cellular biology, molecular biology, biochemistry, and biotechnology. It integrates these disciplines to explore things like genetic inheritance, gene regulation and expression, and the molecular mechanism behind various life processes. A key goal of molecular genetics is to identify and study genetic mutations. Researchers search for mutations in a gene or induce mutations in a gene to link a gene sequence to a specific phenotype. Therefore molecular genetics is a powerful methodology for linking mutations to genetic conditions that may aid th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SeSaM-Biotech GmbH
SeSaM-Biotech GmbH (in short SeSaM-Biotech) is a biotechnology service company founded in 2008 in Bremen and localized in Aachen today. The company has a main focus on Protein Engineering projects based on the QuESt (Quality Enzyme Solutions) strategy and gene library generation using its patented platform technology Sequence Saturation Mutagenesis. Company The SeSaM-Biotech GmbH was founded in 2008 as a spin-off of Jacobs University Bremen Constructor University, formerly Jacobs University Bremen, is an international, private, residential research university located in Vegesack, Bremen, Germany. It offers study programs in engineering, humanities, natural and social sciences, in ... by Prof. Ulrich Schwaneberg who had developed and improved the patented key technology Sequence Saturation Mutagenesis (SeSaM) in the preceding years, In 2012, SeSaM-Biotech moved to its new facilities at the Leibniz-Institute for Interactive Materials (DWI) in Aachen. Today, it is a small co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Terminal Deoxynucleotidyl Transferase
Terminal deoxynucleotidyl transferase (TdT), also known as DNA nucleotidylexotransferase (DNTT) or terminal transferase, is a specialized DNA polymerase expressed in immature, pre-B, pre-T lymphoid cells, and acute lymphoblastic leukemia/lymphoma cells. TdT adds N-nucleotides to the V(D)J recombination, V, D, and J exons of the TCR and BCR genes during antibody gene recombination, enabling the phenomenon of junctional diversity. In humans, terminal transferase is encoded by the ''DNTT'' gene. As a member of the X family of DNA polymerase enzymes, it works in conjunction with polymerase λ and polymerase μ, both of which belong to the same X family of polymerase enzymes. The diversity introduced by TdT has played an important role in the evolution of the vertebrate immune system, significantly increasing the variety of antigen receptors that a cell is equipped with to fight pathogens. Studies using TdT knockout mice have found drastic reductions (10-fold) in T-cell receptor (TCR) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Saturation Mutagenesis
Sequence saturation mutagenesis (SeSaM) is a chemo-enzymatic random mutagenesis method applied for the directed evolution of proteins and enzymes. It is one of the most common saturation mutagenesis techniques. In four PCR-based reaction steps, phosphorothioate nucleotides are inserted in the gene sequence, cleaved and the resulting fragments elongated by universal or degenerate nucleotides. These nucleotides are then replaced by standard nucleotides, allowing for a broad distribution of nucleic acid mutations spread over the gene sequence with a preference to transversions and with a unique focus on consecutive point mutations, both difficult to generate by other mutagenesis techniques. The technique was developed by Professor Ulrich Schwaneberg at Jacobs University Bremen and RWTH Aachen University. Technology, development and advantages SeSaM has been developed in order to overcome several of the major limitations encountered when working with standard mutagenesis methods bas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fusion Protein
Fusion proteins or chimeric (kī-ˈmir-ik) proteins (literally, made of parts from different sources) are proteins created through the joining of two or more genes that originally coded for separate proteins. Translation of this '' fusion gene'' results in a single or multiple polypeptides with functional properties derived from each of the original proteins. ''Recombinant fusion proteins'' are created artificially by recombinant DNA technology for use in biological research or therapeutics. '' Chimeric'' or ''chimera'' usually designate hybrid proteins made of polypeptides having different functions or physico-chemical patterns. ''Chimeric mutant proteins'' occur naturally when a complex mutation, such as a chromosomal translocation, tandem duplication, or retrotransposition creates a novel coding sequence containing parts of the coding sequences from two different genes. Naturally occurring fusion proteins are commonly found in cancer cells, where they may function as oncop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Inosine
Inosine is a nucleoside that is formed when hypoxanthine is attached to a ribose ring (also known as a ribofuranose) via a β-N9-glycosidic bond. It was discovered in 1965 in analysis of RNA transferase. Inosine is commonly found in tRNAs and is essential for proper translation of the genetic code in wobble base pairs. Knowledge of inosine metabolism has led to advances in immunotherapy in recent decades. Inosine monophosphate is oxidised by the enzyme inosine monophosphate dehydrogenase, yielding xanthosine monophosphate, a key precursor in purine metabolism. Mycophenolate mofetil is an anti-metabolite, anti-proliferative drug that acts as an inhibitor of inosine monophosphate dehydrogenase. It is used in the treatment of a variety of autoimmune diseases including granulomatosis with polyangiitis because the uptake of purine by actively dividing B cells can exceed 8 times that of normal body cells, and, therefore, this set of white cells (which cannot operate purine salvage ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid Substitution Pattern In EpPCR And SeSaM
In chemistry, amines (, ) are organic compounds that contain carbon-nitrogen bonds. Amines are formed when one or more hydrogen atoms in ammonia are replaced by alkyl or aryl groups. The nitrogen atom in an amine possesses a lone pair of electrons. Amines can also exist as hetero cyclic compounds. Aniline is the simplest aromatic amine, consisting of a benzene ring bonded to an amino group. Amines are classified into three types: primary (1°), secondary (2°), and tertiary (3°) amines. Primary amines (1°) contain one alkyl or aryl substituent and have the general formula RNH2. Secondary amines (2°) have two alkyl or aryl groups attached to the nitrogen atom, with the general formula R2NH. Tertiary amines (3°) contain three substituent groups bonded to the nitrogen atom, and are represented by the formula R3N. The functional group present in primary amines is called the amino group. Classification of amines Amines can be classified according to the nature and number o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrophobicity
In chemistry, hydrophobicity is the chemical property of a molecule (called a hydrophobe) that is seemingly intermolecular force, repelled from a mass of water. In contrast, hydrophiles are attracted to water. Hydrophobic molecules tend to be chemical polarity#Nonpolar molecules, nonpolar and, thus, prefer other neutral molecules and nonpolar solvents. Because water molecules are polar, hydrophobes do not dissolution (chemistry), dissolve well among them. Hydrophobic molecules in water often cluster together, forming micelles. Water on hydrophobic surfaces will exhibit a high contact angle. Examples of hydrophobic molecules include the alkanes, oils, fats, and greasy substances in general. Hydrophobic materials are used for oil removal from water, the management of oil spills, and chemical separation processes to remove non-polar substances from polar compounds. The term ''hydrophobic''—which comes from the Ancient Greek (), "having a fear of water", constructed Liddell, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conservative Mutation
A conservative replacement (also called a conservative mutation or a conservative substitution or a homologous replacement) is an amino acid replacement in a protein that changes a given amino acid to a different amino acid with similar biochemical properties (e.g. charge, hydrophobicity and size). Conversely, a radical replacement, or radical substitution, is an amino acid replacement that exchanges an initial amino acid by a final amino acid with different physicochemical properties. Description There are 20 naturally occurring amino acids, however some of these share similar characteristics. For example, leucine and isoleucine are both aliphatic, branched hydrophobes. Similarly, aspartic acid and glutamic acid are both small, negatively charged residues. Although there are many ways to classify amino acids, they are often sorted into six main classes on the basis of their structure and the general chemical characteristics of their side chains (R groups). Physicochem ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Synonymous Substitutions
A synonymous substitution (often called a ''silent'' substitution though they are not always silent) is the evolutionary substitution of one base for another in an exon of a gene coding for a protein, such that the produced amino acid sequence is not modified. This is possible because the genetic code is " degenerate", meaning that some amino acids are coded for by more than one three-base-pair codon; since some of the codons for a given amino acid differ by just one base pair from others coding for the same amino acid, a mutation that replaces the "normal" base by one of the alternatives will result in incorporation of the same amino acid into the growing polypeptide chain when the gene is translated. Synonymous substitutions and mutations affecting noncoding DNA are often considered silent mutations; however, it is not always the case that the mutation is silent. Since there are 22 codes for 64 codons, roughly we should expect a random substitution to be synonymous with probabi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |