|
NOD-like Receptor
The nucleotide-binding oligomerization domain-like receptors, or NOD-like receptors (NLRs) (also known as nucleotide-binding leucine-rich repeat receptors), are intracellular sensors of pathogen-associated molecular patterns (PAMPs) that enter the cell via phagocytosis or pores, and damage-associated molecular patterns (DAMPs) that are associated with cell stress. They are types of pattern recognition receptors (PRRs), and play key roles in the regulation of innate immune response. NLRs can cooperate with toll-like receptors (TLRs) and regulate inflammatory and apoptotic response. NLRs primarily recognize Gram-positive bacteria, whereas TLRs primarily recognize Gram-negative bacteria. They are found in lymphocytes, macrophages, dendritic cells and also in non-immune cells, for example in epithelium. NLRs are highly conserved through evolution. Their homologs have been discovered in many different animal species ( APAF1) and also in the plant kingdom ( disease-resistance R pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Leucine-rich Repeat
A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe tertiary structure, fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These Protein tandem repeats, tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta sheet, beta strand-turn (biochemistry), turn-alpha helix structure, and the assembled structural domain, domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues. Leucine-rich repeats are frequently involved in the formation of protein–pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NLRP2
NACHT, LRR and PYD domains-containing protein 2 is a protein that in humans is encoded by the ''NLRP2'' gene. NALP proteins, such as NALP2, are characterized by an N-terminal pyrin domain (PYD) and are involved in the activation of caspase-1 (CASP1; MIM 147678) by Toll-like receptors(see TLR4). They may also be involved in protein complexes that activate proinflammatory caspases (Tschopp et al., 2003). upplied by OMIMref name="entrez"> Function The NLRP2 gene is one of the family members of nucleotide-binding and leucine-rich repeat receptor (NLR). Information from many literature sources indicates that an N-terminal pyrin effector domain (PYD) is one of the components of the NLRP2 gene. Other components include a centrally-located nucleotide-binding and oligomerization domain (NACHT) and C-terminal leucine-rich repeats (LRR). The products of NLRP2 gene are known to interact with IkB kinase (IKK) complex components. It can also regulate the activities of both caspase-1 and nuc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
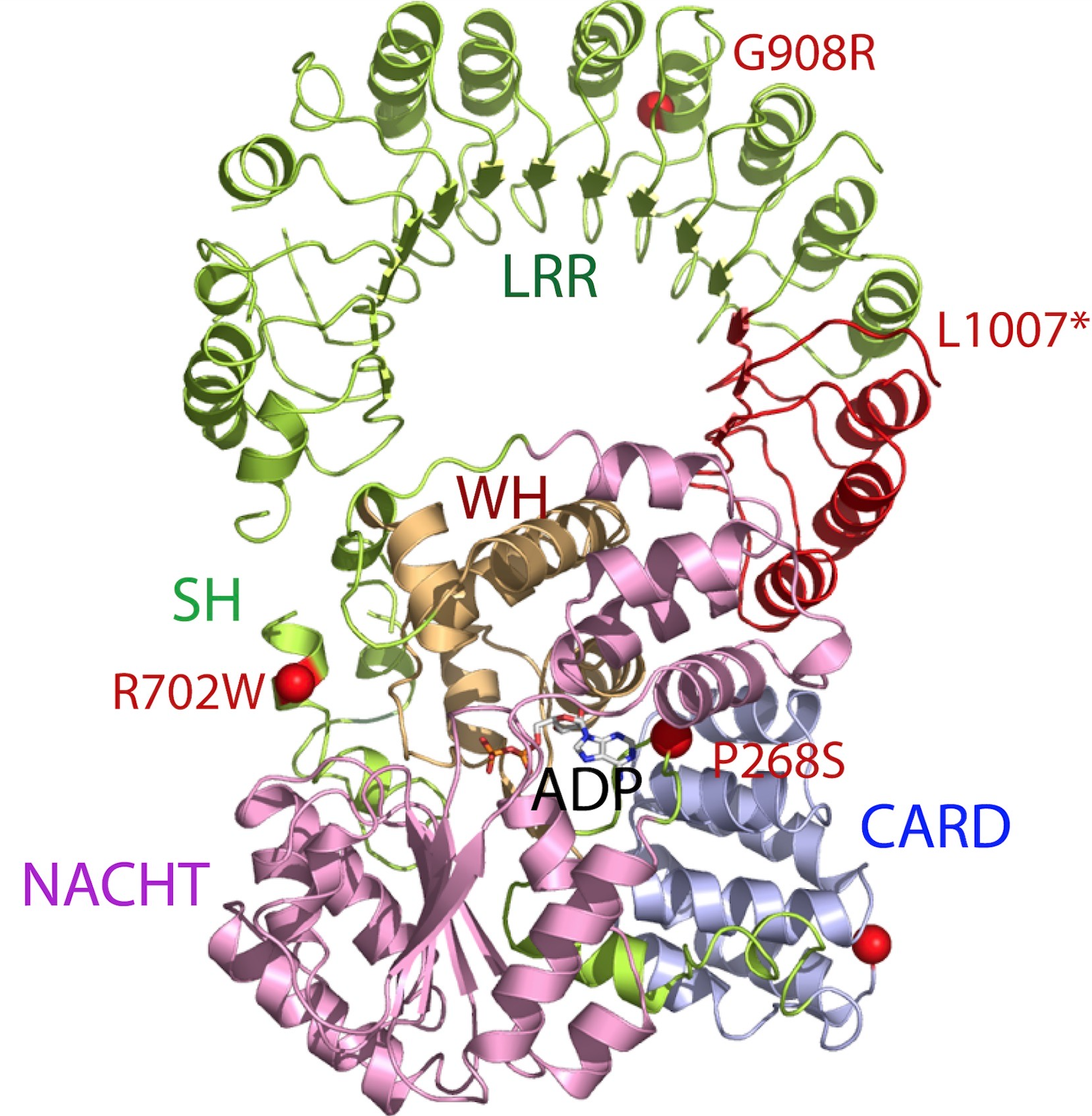
NLRP1
NLRP1 encodes NACHT, LRR, FIIND, CARD domain and PYD domains-containing protein 1 in humans. NLRP1 was the first protein shown to form an inflammasome. Material was copied from this source, which is available under Creative Commons Attribution 4.0 International License NLRP1 is expressed by a variety of cell types, which are predominantly epithelial or hematopoietic. The expression is also seen within glandular epithelial structures including the lining of the small intestine, stomach, airway epithelia and in hairless or glabrous skin. NLRP1 polymorphisms are associated with skin extra-intestinal manifestations in CD. Its highest expression was detected in human skin, in psoriasis and in vitiligo. Polymorphisms of NLRP1 were found in lupus erythematosus and diabetes type 1. Variants of mouse NLRP1 were found to be activated upon N-terminal cleavage by the protease in anthrax lethal factor. Function This gene encodes a member of the Ced-4 family of apoptosis proteins. Ced-fa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NLRC5
NLRC5, short for NOD-like receptor family CARD domain containing 5, is an intracellular protein that plays a role in the immune system. NLRC5 is a pattern recognition receptor implicated in innate immunity to viruses potentially by regulating interferon activity. It also acts as an innate immune sensor to drive inflammatory cell death, PANoptosis. In humans, the NLRC5 protein is encoded by the ''NLRC5'' gene. It has also been called NOD27, NOD4, and CLR16.1. Structure Structurally, NLRC5 has a putative caspase recruitment domain (CARD), followed by a NACHT domain, and a C-terminal leucine-rich repeat ( LRR) region. Function Through its structural features, NLRC5 acts as a key regulator of Major Histocompatibility Class I (MHCI) molecule expression, playing a significant role in the adaptive immune system. This aspect of NLRC5 function was further investigated with the help of ''Nlrc5''- deficient mice, which showed reduced MHCI expression in lymphocytes (particularly T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NLRC4
NLR family CARD domain-containing protein 4 is a protein that in humans is encoded by the ''NLRC4'' gene. Structure The NLRC4 protein is highly conserved across mammalian species. It bears homology to the ''C. elegans'' Ced4 protein. It contains an N-terminal CARD domain, a central nucleotide binding/ NACHT domain, and a C-terminal leucine rich repeat ( LRR) domain. It belongs to a family of NLR proteins that includes the transcriptional co-activator CIITA and the canonical inflammasome protein NLRP3. A truncated murine NLRC4 was the first member of this family whose crystal structure was solved. Function NLRC4 is best associated with triggering formation of the inflammasome. Unlike NLRP3, certain inflammasome-dependent functions of NLRC4 may be carried out independently of the inflammasome scaffold ASC. Human Ced4 homologs include APAF1, NOD1 (CARD4), and NOD2 (CARD15). These proteins have at least 1 N-terminal CARD domain followed by a centrally located nucleotid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NLRC3
NLRC3, short for NOD-like receptor family CARD domain containing 3, is an intracellular protein that plays a role in the immune system. It was previously known as ''nucleotide-binding oligomerization domain, leucine rich repeat and CARD domain containing 3'' (NOD3) and CLR16.2. NLRC3 inhibits the activity of T cells. NLRC3 also inhibits the mTOR The mammalian target of rapamycin (mTOR), also referred to as the mechanistic target of rapamycin, and sometimes called FK506-binding protein 12-rapamycin-associated protein 1 (FRAP1), is a kinase that in humans is encoded by the ''MTOR'' gene. ... signalling pathway to block cellular proliferation. References LRR proteins NOD-like receptors {{Transmembranereceptor-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NOD2
Nucleotide-binding oligomerization domain-containing protein 2 (NOD2), also known as caspase recruitment domain-containing protein 15 (CARD15) or inflammatory bowel disease protein 1 (IBD1), is a protein that in humans is encoded by the ''NOD2'' gene located on chromosome 16. NOD2 plays an important role in the immune system. It recognizes bacterial molecules (peptidoglycans) and stimulates an immune reaction. NOD2 is an intracellular pattern recognition receptor, which is similar in structure to resistant proteins of plants and recognizes molecules containing the specific structure called muramyl dipeptide (MDP) that is found in certain bacteria. Structure The C-terminal portion of the protein contains a leucine-rich repeat domain that is known to play a role in protein–protein interactions. The middle part of the protein is characterized by a NOD domain involved in protein self-oligomerization. The N-terminal portion contains two CARD domains known to play a role in apo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NOD1
Nucleotide-binding oligomerization domain-containing protein 1 (NOD1) is a protein receptor that in humans is encoded by the ''NOD1'' gene. It recognizes bacterial molecules and stimulates an immune reaction. NOD1 protein contains a caspase recruitment domain (CARD). NOD1 is a member of NOD-like receptor protein family and is a close relative of NOD2. NOD1 is an intracellular pattern recognition receptor, which is similar in structure to resistant proteins of plants, and mediates innate and acquired immunity by recognizing molecules containing D-glutamyl-meso-diaminopimelic acid (iE-DAP) moiety, including bacterial peptidoglycan. Nod1 interacts with RIPK2 through the CARDs of both molecules (See the structure of the NOD1 CARD in the right panel). Stimulation of NOD1 by iE-DAP containing molecules results in activation of the transcription factor NF-κB Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) is a family of transcription factor protein complexes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NAIP (gene)
Baculoviral IAP repeat-containing protein 1 is a protein that in humans is encoded by the ''NAIP'' gene. This gene is part of a 500 kb inverted duplication on chromosome 5q13. This duplicated region contains at least four genes and repetitive elements which make it prone to rearrangements and deletions. The repetitiveness and complexity of the sequence have also caused difficulty in determining the organization of this genomic region. This copy of the gene is full length; additional copies with truncations and internal deletions are also present in this region of chromosome 5q13. It is thought that this gene is a modifier of spinal muscular atrophy caused by mutations in a neighboring gene, SMN1. The protein encoded by this gene contains regions of homology to two baculovirus inhibitor of apoptosis Apoptosis (from ) is a form of programmed cell death that occurs in multicellular organisms and in some eukaryotic, single-celled microorganisms such as yeast. Biochemistry, Bioch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CIITA
CIITA is a human gene which encodes a protein called the class II, major histocompatibility complex, transactivator. Mutations in this gene are responsible for the bare lymphocyte syndrome in which the immune system is severely compromised and cannot effectively fight infection. Chromosomal rearrangement of CIITA is involved in the pathogenesis of Hodgkin lymphoma and primary mediastinal B cell lymphoma. Function CIITA mRNA can only be detected in human leukocyte antigen (HLA) system class II-positive cell lines and tissues. This highly restricted tissue distribution suggests that expression of HLA class II genes is to a large extent under the control of CIITA. However, CIITA does not appear to directly bind to DNA. Instead CIITA functions through activation of the transcription factor RFX5. Hence CIITA is classified as a transcriptional coactivator. The CIITA protein contains an acidic transcriptional activation domain, 4 LRRs ( leucine-rich repeats) and a GTP binding d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HUGO Gene Nomenclature Committee
The HUGO Gene Nomenclature Committee (HGNC) is a committee of the Human Genome Organisation (HUGO) that sets the standards for human gene nomenclature. The HGNC approves a ''unique'' and ''meaningful'' name for every known human gene, based on a query of experts. In addition to the name, which is usually 1 to 10 words long, the HGNC also assigns a symbol (a short group of characters) to every gene. As with an SI symbol, a gene symbol is like an abbreviation but is more than that, being a second unique name that can stand on its own just as much as substitute for the longer name. It may not necessarily "stand for" the initials of the name, although many gene symbols do reflect that origin. Purpose Full gene names, and especially gene abbreviations and symbols, are often not specific to a single gene. A marked example is CAP which can refer to any of 6 different genes (BRD4'', CAP1'', HACD1'', LNPEP'', SERPINB6'', and SORBS1''). The HGNC short gene names, or gene symbols, unlik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |