|
LTR Retrotransposons
LTR retrotransposons are class I transposable element characterized by the presence of long terminal repeats (LTRs) directly flanking an internal coding region. As retrotransposons, they mobilize through reverse transcription of their mRNA and integration of the newly created cDNA into another location. Their mechanism of retrotransposition is shared with retroviruses, with the difference that most LTR-retrotransposons do not form infectious particles that leave the cells and therefore only replicate inside their genome of origin. Those that do (occasionally) form virus-like particles are classified under ''Ortervirales''. Their size ranges from a few hundred base pairs to 25kb, for example the Ogre retrotransposon in the pea genome. In plant genomes, LTR retrotransposons are the major repetitive sequence class, for example, constituting more than 75% of the maize genome. LTR retrotransposons make up about 8% of the human genome and approximately 10% of the mouse genome. Struc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LTR Retrotransposon
LTR retrotransposons are class I transposable element characterized by the presence of long terminal repeats (LTRs) directly flanking an internal coding region. As retrotransposons, they mobilize through reverse transcription of their mRNA and integration of the newly created cDNA into another location. Their mechanism of retrotransposition is shared with retroviruses, with the difference that most LTR-retrotransposons do not form infectious particles that leave the cells and therefore only replicate inside their genome of origin. Those that do (occasionally) form virus-like particles are classified under ''Ortervirales''. Their size ranges from a few hundred base pairs to 25kb, for example the Ogre retrotransposon in the pea genome. In plant genomes, LTR retrotransposons are the major repetitive sequence class, for example, constituting more than 75% of the maize genome. LTR retrotransposons make up about 8% of the human genome and approximately 10% of the mouse genome. Struc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Hierarchy of proteases Based on catalytic residue Proteases can be classified into seven broad groups: * Serine protea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
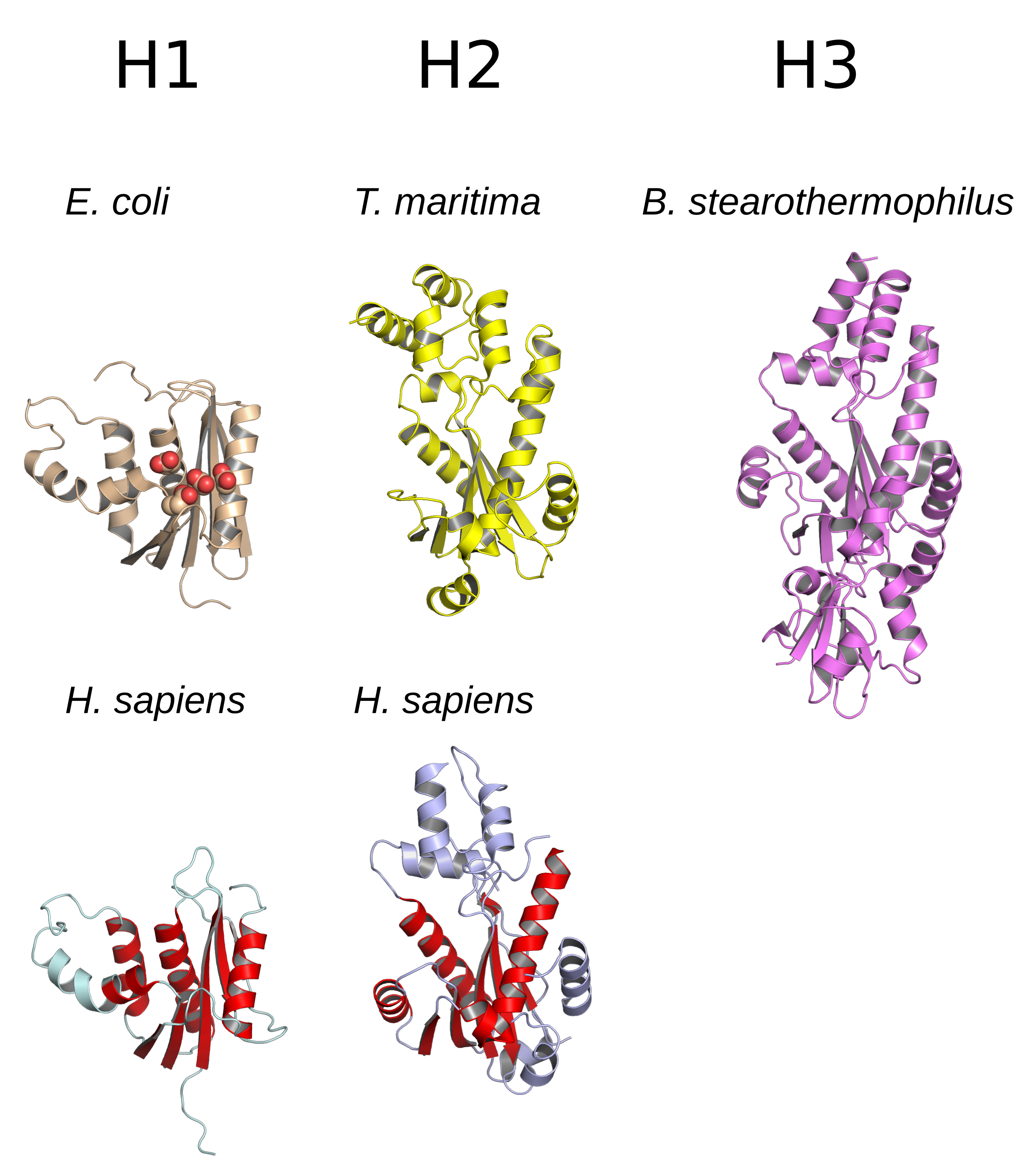
Ribonuclease H
Ribonuclease H (abbreviated RNase H or RNH) is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/ DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes. The family is divided into evolutionarily related groups with slightly different substrate preferences, broadly designated ribonuclease H1 and H2. The human genome encodes both H1 and H2. Human ribonuclease H2 is a heterotrimeric complex composed of three subunits, mutations in any of which are among the genetic causes of a rare disease known as Aicardi–Goutières syndrome. A third type, closely related to H2, is found only in a few prokaryotes, whereas H1 and H2 occur in all domains of life. Additionally, RNase H1-like retroviral ribonuclease H domains occur in multidomain reverse transcriptase proteins, which are encoded by retroviruses such as HIV and are required for vira ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Angiosperms
Flowering plants are plants that bear flowers and fruits, and form the clade Angiospermae (), commonly called angiosperms. The term "angiosperm" is derived from the Greek words ('container, vessel') and ('seed'), and refers to those plants that produce their seeds enclosed within a fruit. They are by far the most diverse group of land plants with 64 orders, 416 families, approximately 13,000 known genera and 300,000 known species. Angiosperms were formerly called Magnoliophyta (). Like gymnosperms, angiosperms are seed-producing plants. They are distinguished from gymnosperms by characteristics including flowers, endosperm within their seeds, and the production of fruits that contain the seeds. The ancestors of flowering plants diverged from the common ancestor of all living gymnosperms before the end of the Carboniferous, over 300 million years ago. The closest fossil relatives of flowering plants are uncertain and contentious. The earliest angiosperm fossils are in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gymnosperms
The gymnosperms ( lit. revealed seeds) are a group of seed-producing plants that includes conifers, cycads, '' Ginkgo'', and gnetophytes, forming the clade Gymnospermae. The term ''gymnosperm'' comes from the composite word in el, γυμνόσπερμος ( el, γυμνός, translit=gymnos, lit=naked, label=none and el, σπέρμα, translit=sperma, lit=seed, label=none), literally meaning 'naked seeds'. The name is based on the unenclosed condition of their seeds (called ovules in their unfertilized state). The non-encased condition of their seeds contrasts with the seeds and ovules of flowering plants (angiosperms), which are enclosed within an ovary. Gymnosperm seeds develop either on the surface of scales or leaves, which are often modified to form cones, or solitary as in yew, '' Torreya'', '' Ginkgo''. Gymnosperm lifecycles involve alternation of generations. They have a dominant diploid sporophyte phase and a reduced haploid gametophyte phase which is dependent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bryophytes
The Bryophyta s.l. are a proposed taxonomic division containing three groups of non-vascular land plants ( embryophytes): the liverworts, hornworts and mosses. Bryophyta s.s. consists of the mosses only. They are characteristically limited in size and prefer moist habitats although they can survive in drier environments. The bryophytes consist of about 20,000 plant species. Bryophytes produce enclosed reproductive structures (gametangia and sporangia), but they do not produce flowers or seeds. They reproduce sexually by spores and asexually by fragmentation or the production of gemmae. Though bryophytes were considered a paraphyletic group in recent years, almost all of the most recent phylogenetic evidence supports the monophyly of this group, as originally classified by Wilhelm Schimper in 1879. The term ''bryophyte'' comes . Terminology The term "Bryophyta" was first suggested by Braun in 1864. G.M. Smith placed this group between Algae and Pteridophyta. Feature ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Algae
Algae (; singular alga ) is an informal term for a large and diverse group of photosynthetic eukaryotic organisms. It is a polyphyletic grouping that includes species from multiple distinct clades. Included organisms range from unicellular microalgae, such as ''Chlorella,'' ''Prototheca'' and the diatoms, to multicellular forms, such as the giant kelp, a large brown alga which may grow up to in length. Most are aquatic and autotrophic (they generate food internally) and lack many of the distinct cell and tissue types, such as stomata, xylem and phloem that are found in land plants. The largest and most complex marine algae are called seaweeds, while the most complex freshwater forms are the ''Charophyta'', a division of green algae which includes, for example, ''Spirogyra'' and stoneworts. No definition of algae is generally accepted. One definition is that algae "have chlorophyll ''a'' as their primary photosynthetic pigment and lack a sterile covering of cells around their re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRNAs
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino acid sequence of proteins. tRNAs genes from Bacteria are typically shorter (mean = 77.6 bp) than tRNAs from Archaea (mean = 83.1 bp) and eukaryotes (mean = 84.7 bp). The mature tRNA follows an opposite pattern with tRNAs from Bacteria being usually longer (median = 77.6 nt) than tRNAs from Archaea (median = 76.8 nt), with eukaryotes exhibiting the shortest mature tRNAs (median = 74.5 nt). Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
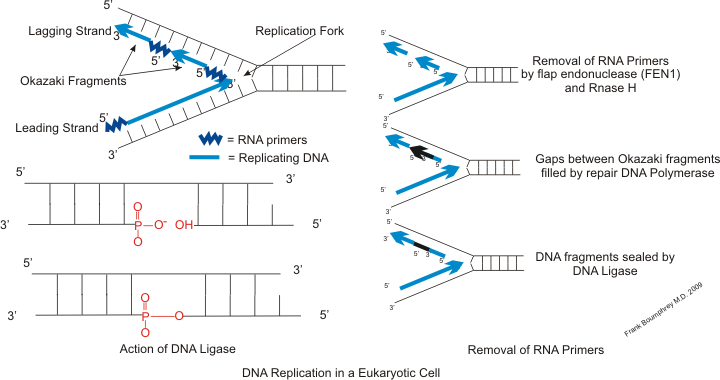
Primer Binding Site
A primer binding site is a region of a nucleotide sequence where an RNA or DNA single-stranded primer binds to start replication. The primer binding site is on one of the two complementary strands of a double-stranded nucleotide polymer, in the strand which is to be copied, or is within a single-stranded nucleotide polymer sequence. DNA Replication DNA replication is the semi-conservative, biological process of two DNA strands copying themselves, resulting in two identical copies of DNA. This process is considered semi-conservative because, after replication, each copy of DNA contains a strand from the original DNA molecule and a strand from the newly-synthesized DNA molecule. An RNA primer is a short chain of single-stranded RNA, consisting of roughly five to ten nucleotides complementary to the DNA template strand. DNA polymerase will then take each nucleotide and make a new complementary DNA strand to the template strand, but only in the 5' to 3' direction. One of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcription
A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible. Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. Al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase II
RNA polymerase II (RNAP II and Pol II) is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoters and begin transcription. Discovery Early studies suggested a minimum of two RNAPs: one which synthesized rRNA in the nucleolus, and one which synthesized other RNA in the nucleoplasm, part of the nucleus but outside the nucleolus. In 1969, science experimentalists Robert Roeder and William Rutter definitively discovered an additional RNAP that was responsible for transcription of some kind of RNA in the nucleoplasm. The finding was obtained by the use of ion-exchange chromatography via DEAE coated Sephadex beads. The technique separated the enz ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |