|
KCTD7
Potassium channel tetramerisation domain containing 7 is a protein in humans that is encoded by the KCTD7 gene. Alternative splicing results in multiple transcript variants. Description The KCTD7 gene encodes a member of the potassium channel tetramerisation domain-containing protein family. Family members are identified on a structural basis and contain an amino-terminal domain similar to the T1 domain present in the voltage-gated potassium channel. KCTD7 displays a primary sequence and hydropathy profile indicating intracytoplasmic localization. EST database analysis showed that KCTD7 is expressed in human and mouse brain. Function KCTD7 expression hyperpolarizes the cell membrane and reduces the excitability of transfected neurons in patch clamp experiments. KCTD7 mRNA and protein are expressed in hippocampal neurons, deep layers of the cerebral cortex and Purkinje cells of the murine brain as shown by in situ hybridization and immunohistochemistry experiments. Immunopr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Potassium Channel Tetramerisation Domain
K+ channel tetramerisation domain is the N-terminal, cytoplasmic tetramerisation domain (T1) of voltage-gated K+ channels. It defines molecular determinants for subfamily-specific assembly of alpha-subunits into functional tetrameric channels. It is distantly related to the BTB/POZ domain . Potassium channels Potassium channels are the most diverse group of the ion channel family. They are important in shaping the action potential, and in neuronal excitability and plasticity. The potassium channel family is composed of several functionally distinct isoforms, which can be broadly separated into 2 groups: the practically non-inactivating 'delayed' group and the rapidly inactivating 'transient' group. These are all highly similar proteins, with only small amino acid changes causing the diversity of the voltage-dependent gating mechanism, channel conductance and toxin binding properties. Each type of K+ channel is activated by different signals and conditions depending on their t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid resid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
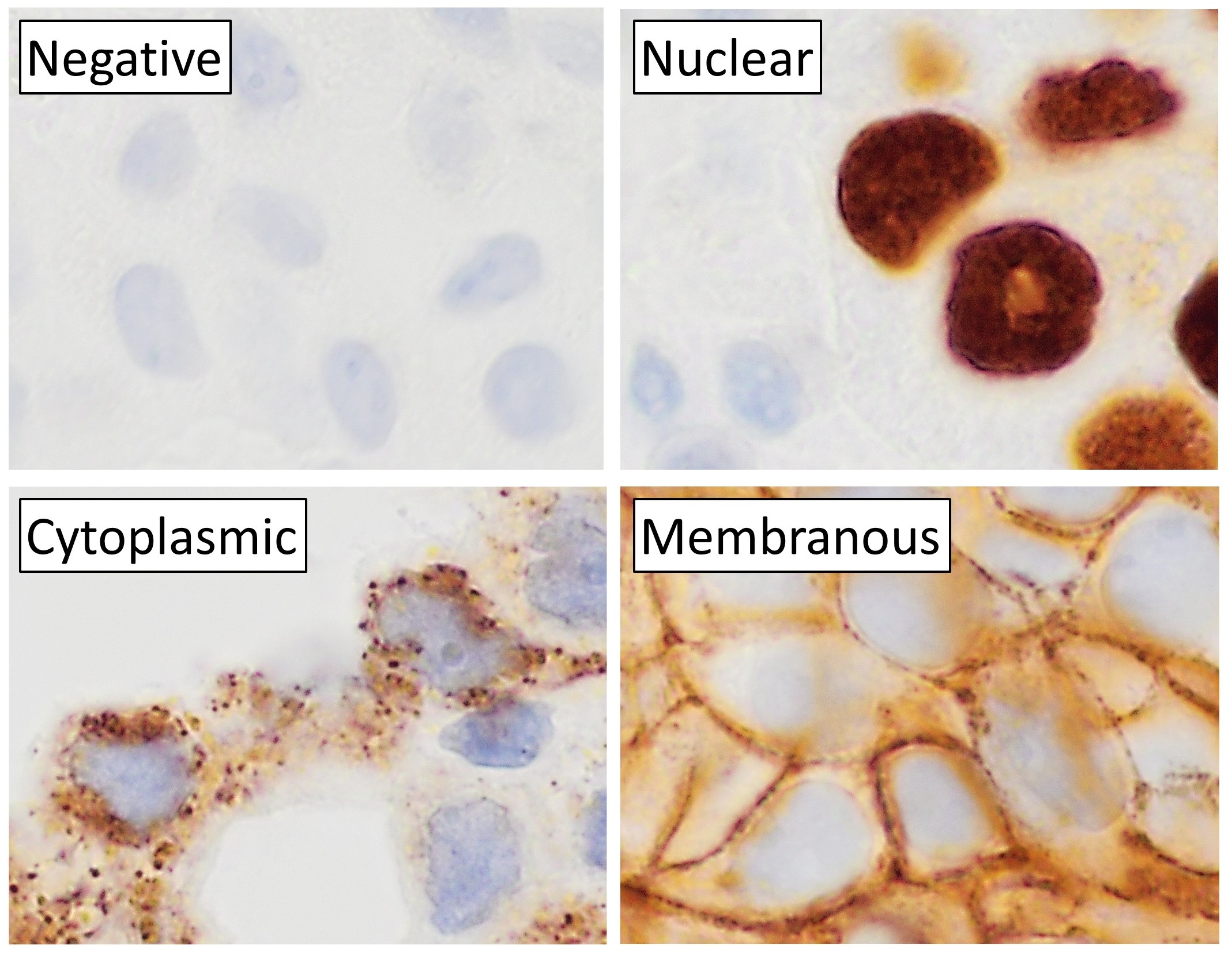
Immunohistochemistry
Immunohistochemistry (IHC) is the most common application of immunostaining. It involves the process of selectively identifying antigens (proteins) in cells of a tissue section by exploiting the principle of antibodies binding specifically to antigens in biological tissues. IHC takes its name from the roots "immuno", in reference to antibodies used in the procedure, and "histo", meaning tissue (compare to immunocytochemistry). Albert Coons conceptualized and first implemented the procedure in 1941. Visualising an antibody-antigen interaction can be accomplished in a number of ways, mainly either of the following: * ''Chromogenic immunohistochemistry'' (CIH), wherein an antibody is conjugated to an enzyme, such as peroxidase (the combination being termed immunoperoxidase), that can catalyse a colour-producing reaction. * '' Immunofluorescence'', where the antibody is tagged to a fluorophore, such as fluorescein or rhodamine. Immunohistochemical staining is widely used in t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Myoclonic Epilepsy
Myoclonic epilepsy refers to a family of epilepsies that present with myoclonus. It starts in both sides of the body at once, and last for more than a second or two. When myoclonic jerks are occasionally associated with abnormal brain wave activity, it can be categorized as myoclonic seizure. If the abnormal brain wave activity is persistent and results from ongoing seizures, then a diagnosis of myoclonic epilepsy may be considered. Myoclonic seizures frequently occur in day-to-day life. During sleep, abrupt jerks and hiccups occurred. Myoclonic jerks are typical without regularity. Myoclonus can be a source of substantial disability with impairment in daily activities. Myoclonus can be termed positive or negative, A sudden muscle contraction is known as "positive myoclonus," and a brief loss of muscular tone is known as "negative myoclonus". Myoclonus which occurred at rest indicates the source as a spinal or brainstem. Myoclonus which is action induced indicates the source as c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin Ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another thing (the substrate) by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Potassium Channel
Potassium channels are the most widely distributed type of ion channel found in virtually all organisms. They form potassium-selective pores that span cell membranes. Potassium channels are found in most cell types and control a wide variety of cell functions. Function Potassium channels function to conduct potassium ions down their electrochemical gradient, doing so both rapidly (up to the diffusion rate of K+ ions in bulk water) and selectively (excluding, most notably, sodium despite the sub-angstrom difference in ionic radius). Biologically, these channels act to set or reset the resting potential in many cells. In excitable cells, such as neurons, the delayed counterflow of potassium ions shapes the action potential. By contributing to the regulation of the cardiac action potential duration in cardiac muscle, malfunction of potassium channels may cause life-threatening arrhythmias. Potassium channels may also be involved in maintaining vascular tone. They also re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
COS Cells
COS are fibroblast-like cell lines derived from monkey kidney tissue. COS cells are obtained by immortalizing CV-1 cells with a version of the SV40 virus that can produce large T antigen but has a defect in genomic replication. The CV-1 cell line in turn was derived from the kidney of the African green monkey. The acronym "COS" is derived from the cells being CV-1 (simian) in Origin, and carrying the SV40 genetic material. Three COS lines were created (COS-1, COS-3 and COS-7), of which two are commonly used (COS-1 and COS-7). Applications The COS cell lines are often used by biologists when studying the monkey virus SV40. Cells from these lines are also often transfected to produce recombinant proteins for molecular biology, biochemistry, and cell biology experiments. When an expression construct with an SV40 origin of replication is introduced into COS cells, the vector can be replicated substantially by the large T antigen The large tumor antigen (also called the large T-an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Green Fluorescent Protein
The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range. The label ''GFP'' traditionally refers to the protein first isolated from the jellyfish '' Aequorea victoria'' and is sometimes called ''avGFP''. However, GFPs have been found in other organisms including corals, sea anemones, zoanithids, copepods and lancelets. The GFP from ''A. victoria'' has a major excitation peak at a wavelength of 395 nm and a minor one at 475 nm. Its emission peak is at 509 nm, which is in the lower green portion of the visible spectrum. The fluorescence quantum yield (QY) of GFP is 0.79. The GFP from the sea pansy (''Renilla reniformis'') has a single major excitation peak at 498 nm. GFP makes for an excellent tool in many forms of biology due to its ability to form an internal chromophore without requiring any accessory cofactors, gene products, or enzymes / substrates other tha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
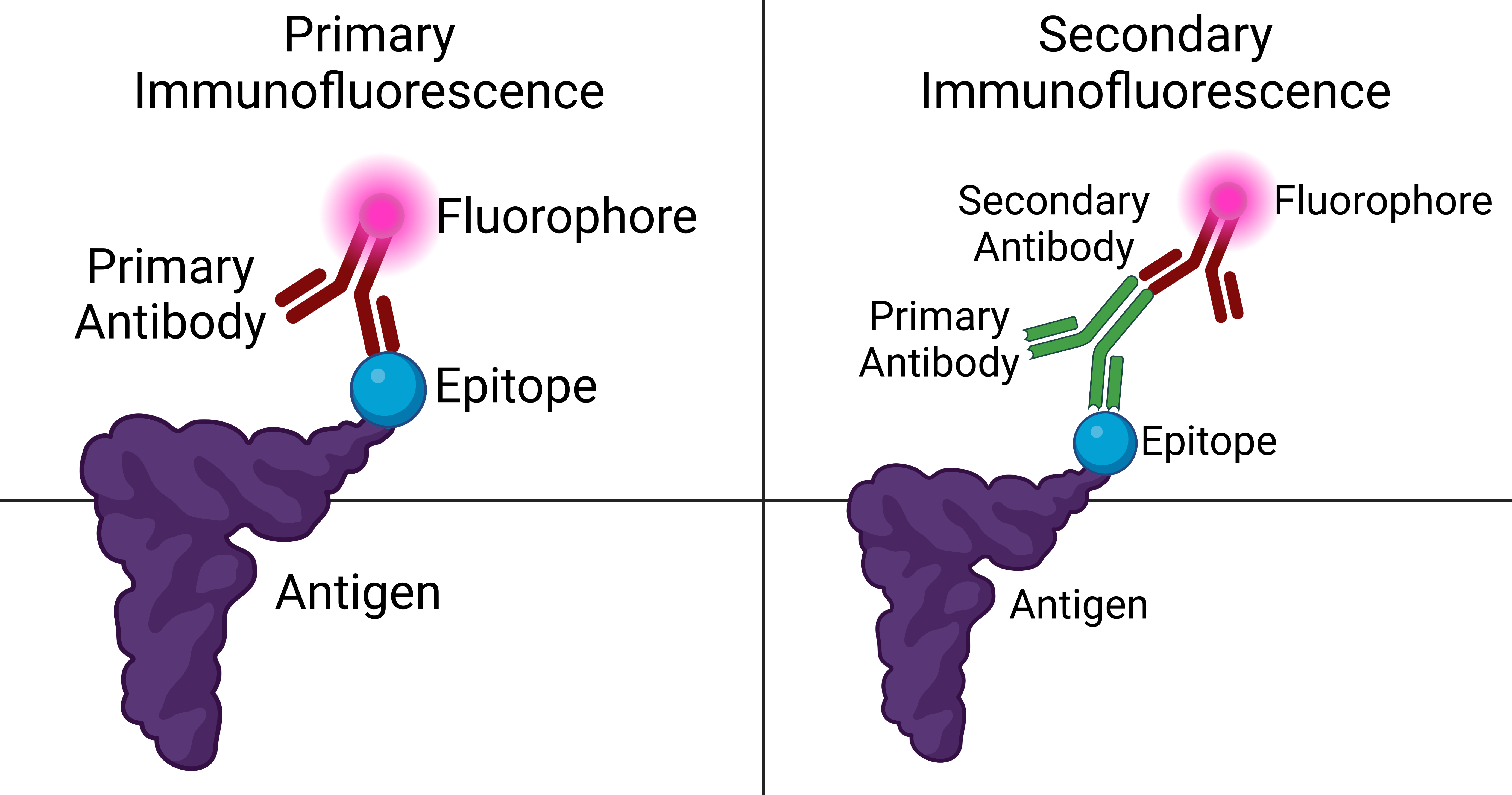
Immunofluorescence Microscopy
Immunofluorescence is a technique used for light microscopy with a fluorescence microscope and is used primarily on microbiological samples. This technique uses the specificity of antibodies to their antigen to target fluorescent dyes to specific biomolecule targets within a cell, and therefore allows visualization of the distribution of the target molecule through the sample. The specific region an antibody recognizes on an antigen is called an epitope. There have been efforts in epitope mapping since many antibodies can bind the same epitope and levels of binding between antibodies that recognize the same epitope can vary. Additionally, the binding of the fluorophore to the antibody itself cannot interfere with the immunological specificity of the antibody or the binding capacity of its antigen. Immunofluorescence is a widely used example of immunostaining (using antibodies to stain proteins) and is a specific example of immunohistochemistry (the use of the antibody-antigen re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BTB/POZ Domain
The BTB/POZ domain is a common structural domain contained within some proteins. The BTB (for BR-C, ttk and bab) or POZ (for Pox virus and Zinc finger) domain is present near the N-terminus of a fraction of zinc finger proteins and in proteins that contain the Kelch motif and a family of pox virus proteins. The BTB/POZ domain mediates homomeric dimerisation and in some instances heteromeric dimerisation. The structure of the dimerised PLZF BTB/POZ domain has been solved and consists of a tightly intertwined homodimer. The central scaffolding of the protein is made up of a cluster of alpha-helices flanked by short beta-sheets at both the top and bottom of the molecule. BTB/POZ domains from several zinc finger proteins have been shown to mediate transcriptional repression and to interact with components of histone deacetylase co-repressor complexes including N-CoR The nuclear receptor co-repressor 1 also known as thyroid-hormone- and retinoic-acid-receptor-associated co-repressor 1 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CUL3
Cullin 3 is a protein that in humans is encoded by the ''CUL3'' gene. Cullin 3 protein belongs to the family of cullins which in mammals contains eight proteins (Cullin 1, Cullin 2, Cullin 3, Cullin 4A, Cullin 4B, Cullin 5, Cullin 7 and Cullin 9). Cullin proteins are an evolutionarily conserved family of proteins throughout yeast, plants and mammals. Protein function Cullin 3 is a component of Cullin- RING E3 ubiquitin ligases complexes (CRLs) which are involved in protein ubiquitylation and represent a part of ubiquitin–proteasome system (UPS). Added ubiquitin moieties to the lysine residue by CRLs then target the protein for the proteasomal degradation. Cullin- RING E3 ubiquitin ligases are involved in many cellular processes responsible for cell cycle regulation, stress response, protein trafficking, signal transduction, DNA replication, transcription, protein quality control, circadian clock and development. Deletion of ''CUL3'' gene in mice causes embryonic lethality ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |