|
Heterochromatin
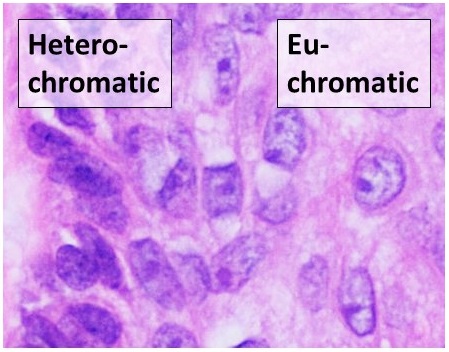
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed; however, according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin. Constitutive heterochromatin can affect the genes near itself (e.g. position-effect variegation). It is usually repetitive and forms structural functions such as centromeres or telomeres, in addition to acting as an attractor for other gene-expression or repression signals. Facultativ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heterochromatin Vs
Heterochromatin is a tightly packed form of DNA or ''DNA_condensation, condensed DNA'', which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the gene expression, expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed; however, according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously Messenger RNA#Eukaryotic mRNA turnover, turned over via RNA-induced transcriptional silencing (RITS). Recent studies with Electron microscope, electron microscopy and osmium tetroxide, OsO4 staining reveal that the dense packing is not due to the chromatin. Constitutive heterochromatin can affect the genes near itself (e.g. position-effect variegation). It is usually Repeated sequence (DNA), repetitive and forms structural functions such ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Facultative Heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed; however, according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin. Constitutive heterochromatin can affect the genes near itself (e.g. position-effect variegation). It is usually repetitive and forms structural functions such as centromeres or telomeres, in addition to acting as an attractor for other gene-expression or repression signals. Facultative ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Constitutive Heterochromatin
Constitutive heterochromatin domains are regions of DNA found throughout the chromosomes of eukaryotes. The majority of constitutive heterochromatin is found at the pericentromeric regions of chromosomes, but is also found at the telomeres and throughout the chromosomes. In humans there is significantly more constitutive heterochromatin found on chromosomes 1, 9, 16, 19 and Y. Constitutive heterochromatin is composed mainly of high copy number tandem repeats known as satellite repeats, minisatellite and microsatellite repeats, and Transposable element, transposon repeats. In humans these regions account for about 200Mb or 6.5% of the total human genome, but their repeat composition makes them difficult to sequence, so only small regions have been sequenced. Visualization of constitutive heterochromatin is possible by using the Cytogenetics, C-banding technique. The regions that stain darker are regions of constitutive heterochromatin. The constitutive heterochromatin stains darker ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Euchromatin
Euchromatin (also called "open chromatin") is a lightly packed form of chromatin (DNA, RNA, and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin stands in contrast to heterochromatin, which is tightly packed and less accessible for transcription. 92% of the human genome is euchromatic. In eukaryotes, euchromatin comprises the most active portion of the genome within the cell nucleus. In prokaryotes, euchromatin is the ''only'' form of chromatin present; this indicates that the heterochromatin structure evolved later along with the nucleus, possibly as a mechanism to handle increasing genome size. Structure Euchromatin is composed of repeating subunits known as nucleosomes, reminiscent of an unfolded set of beads on a string, that are approximately 11 nm in diameter. At the core of these nucleosomes are a set of four histone protein pairs: H3, H4, H2A, and H2B. Each core histone protein possesses a 'tail' structu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-induced Transcriptional Silencing
RNA-induced transcriptional silencing (RITS) is a form of RNA interference by which short RNA molecules – such as small interfering RNA (siRNA) – trigger the downregulation of transcription of a particular gene or genomic region. This is usually accomplished by posttranslational modification of histone tails (e.g. methylation of lysine 9 of histone H3) which target the genomic region for heterochromatin formation. The protein complex that binds to siRNAs and interacts with the methylated lysine 9 residue of histones H3 ( H3K9me2) is the RITS complex. RITS was discovered in the fission yeast ''Schizosaccharomyces pombe'', and has been shown to be involved in the initiation and spreading of heterochromatin in the mating-type region and in centromere formation. The RITS complex in ''S. pombe'' contains at least a piwi domain-containing RNase H-like argonaute, a chromodomain protein Chp1, and an argonaute interacting protein Tas3 which can also bind to Chp1, while heterochrom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetic
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional (DNA sequence based) genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological traits may result from environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Further, non-coding RNA sequences have been shown to play a key role in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H3K9me3
H3K9me3 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the tri-methylation at the 9th lysine residue of the histone H3 protein and is often associated with heterochromatin. Nomenclature H3K9me3 indicates trimethylation of lysine 9 on histone H3 protein subunit: Lysine methylation : This diagram shows the progressive methylation of a lysine residue. The tri-methylation (right) denotes the methylation present in H3K9me3 . Understanding histone modifications The genomic DNA of eukaryotic cells is wrapped around special protein molecules known as Histones. The complexes formed by the looping of the DNA are known as chromatin. The basic structural unit of chromatin is the nucleosome: this consists of the core octamer of histones (H2A, H2B, H3 and H4) as well as a linker histone and about 180 base pairs of DNA. These core histones are rich in lysine and arginine residues. The carboxyl (C) terminal end of these histones contrib ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including ''co-suppression'', ''post-transcriptional gene silencing'' (PTGS), and ''quelling''. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm ''Caenorhabditis elegans'', which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
X Chromosome
The X chromosome is one of the two sex chromosomes in many organisms, including mammals, and is found in both males and females. It is a part of the XY sex-determination system and XO sex-determination system. The X chromosome was named for its unique properties by early researchers, which resulted in the naming of its counterpart Y chromosome, for the next letter in the alphabet, following its subsequent discovery. Discovery It was first noted that the X chromosome was special in 1890 by Hermann Henking in Leipzig. Henking was studying the testicles of '' Pyrrhocoris'' and noticed that one chromosome did not take part in meiosis. Chromosomes are so named because of their ability to take up staining (''chroma'' in Greek means ''color''). Although the X chromosome could be stained just as well as the others, Henking was unsure whether it was a different class of the object and consequently named it ''X element'', which later became X chromosome after it was established that it w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Barr Body
A Barr body (named after discoverer Murray Barr) or X-chromatin is an inactive X chromosome. In species with XY sex-determination (including humans), females typically have two X chromosomes, and one is rendered inactive in a process called lyonization. Errors in chromosome separation can also result in male and female individuals with extra X chromosomes. The Lyon hypothesis states that in cells with multiple X chromosomes, all but one are inactivated early in embryonic development in mammals.Brown, C.J., Robinson, W.P., (1997), XIST Expression and X-Chromosome Inactivation in Human Preimplantation Embryos ''Am. J. Hum. Genet.'' 61, 5–8Full Text PDF The X chromosomes that become inactivated are chosen randomly, except in marsupials and in some extra-embryonic tissues of some placental mammals, in which the X chromosome from the sperm is always deactivated. In humans with euploidy, a genotypical female (46, XX karyotype) has one Barr body per somatic cell nucleus, w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Position-effect Variegation
Position-effect variegation (PEV) is a Variegation (histology), variegation caused by the silencing of a gene in some cells through its abnormal juxtaposition with heterochromatin via rearrangement or Transposable element, transposition. It is also associated with changes in Chromatin, chromatin conformation. Overview The classical example is the ''Drosophila'' wm4 (speak white-mottled-4) Chromosomal translocation, translocation. In this mutation, an Chromosomal inversion, inversion on the X chromosome placed the ''white'' gene next to pericentric heterochromatin, or a sequence of repeats that becomes heterochromatic. Normally, the ''white'' gene is expressed in every cell of the adult ''Drosophila'' eye resulting in a red-eye phenotype. In the w[m4] mutant, the eye color was variegated (red-white mosaic colored) where the ''white'' gene was expressed in some cells in the eyes and not in others. The mutation was described first by Hermann Joseph Muller, Hermann Muller in 1930. PEV ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Repeated Sequence (DNA)
Repeated sequences (also known as repetitive elements, repeating units or repeats) are short or long patterns that occur in multiple copies throughout the genome. In many organisms, a significant fraction of the genomic DNA is repetitive, with over two-thirds of the sequence consisting of repetitive elements in humans. Some of these repeated sequences are necessary for maintaining important genome structures such as telomeres or centromeres. Repeated sequences are categorized into different classes depending on features such as structure, length, location, origin, and mode of multiplication. The disposition of repetitive elements throughout the genome can consist either in directly adjacent arrays called tandem repeats or in repeats dispersed throughout the genome called interspersed repeats. Tandem repeats and interspersed repeats are further categorized into subclasses based on the length of the repeated sequence and/or the mode of multiplication. While some repeated DNA seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |