|
Gene Knockout
Gene knockouts (also known as gene deletion or gene inactivation) are a widely used genetic engineering technique that involves the gene targeting, targeted removal or inactivation of a specific gene within an organism's genome. This can be done through a variety of methods, including homologous recombination, CRISPR gene editing, CRISPR-Cas9, and transcription activator-like effector nuclease, TALENs. One of the main advantages of gene knockouts is that they allow researchers to study the function of a specific gene in vivo, and to understand the role of the gene in normal development and physiology as well as in the pathology of diseases. By studying the phenotype of the organism with the knocked out gene, researchers can gain insights into the biological processes that the gene is involved in. There are two main types of gene knockouts: complete and conditional. A complete gene knockout permanently inactivates the gene, while a conditional gene knockout allows for the gene to b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Targeting
Gene targeting is a biotechnological tool used to change the DNA sequence of an organism (hence it is a form of Genome Editing). It is based on the natural DNA-repair mechanism of Homology Directed Repair (HDR), including Homologous Recombination. Gene targeting can be used to make a range of sizes of DNA edits, from larger DNA edits such as inserting entire new genes into an organism, through to much smaller changes to the existing DNA such as a single base-pair change. Gene targeting relies on the presence of a repair template to introduce the user-defined edits to the DNA. The user (usually a scientist) will design the repair template to contain the desired edit, flanked by DNA sequence corresponding (homologous) to the region of DNA that the user wants to edit; hence the edit is ''targeted'' to a particular genomic region. In this way Gene Targeting is distinct from natural homology-directed repair, during which the ‘natural’ DNA repair template of the sister chromatid i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electroporation
Electroporation, also known as electropermeabilization, is a microbiological and biotechnological technique in which an electric field is applied to cells to briefly increase the permeability of the cell membrane. The application of a high-voltage electric field induces a temporary destabilization of the lipid bilayer, resulting in the formation of nanoscale pores that permit the entry or exit of macromolecules. This method is widely employed to introduce molecules—including small molecules, DNA, RNA, and proteins—into cells. Electroporation can be performed on cells in suspension using electroporation cuvettes, or directly on adherent cells ''in situ'' within their culture vessels. In microbiology, electroporation is frequently utilized for the transformation of bacteria or yeast cells, often with plasmid DNA. It is also used in the transfection of plant protoplasts and mammalian cells. Notably, electroporation plays a critical role in the '' ex vivo'' manipulation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Blood–brain Barrier
The blood–brain barrier (BBB) is a highly selective semipermeable membrane, semipermeable border of endothelium, endothelial cells that regulates the transfer of solutes and chemicals between the circulatory system and the central nervous system, thus protecting the brain from harmful or unwanted substances in the blood. The blood–brain barrier is formed by endothelial cells of the Capillary, capillary wall, astrocyte end-feet ensheathing the capillary, and pericytes embedded in the capillary basement membrane. This system allows the passage of some small molecules by passive transport, passive diffusion, as well as the selective and active transport of various nutrients, ions, organic anions, and macromolecules such as glucose and amino acids that are crucial to neural function. The blood–brain barrier restricts the passage of pathogens, the diffusion of solutes in the blood, and Molecular mass, large or Hydrophile, hydrophilic molecules into the cerebrospinal fluid, while a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TALENs
Transcription activator-like effector nucleases (TALEN) are restriction enzymes that can be engineered to cut specific sequences of DNA. They are made by fusing a TAL effector DNA-binding domain to a DNA cleavage domain (a nuclease which cuts DNA strands). Transcription activator-like effectors (TALEs) can be engineered to bind to practically any desired DNA sequence, so when combined with a nuclease, DNA can be cut at specific locations. The restriction enzymes can be introduced into cells, for use in gene editing or for genome editing ''in situ'', a technique known as genome editing with engineered nucleases. Alongside zinc finger nucleases and CRISPR/Cas9, TALEN is a prominent tool in the field of genome editing. TALE DNA-binding domain TAL effectors are proteins that are secreted by ''Xanthomonas'' bacteria via their type III secretion system when they infect plants. The DNA binding domain contains a repeated highly conserved 33–34 amino acid sequence with divergent 1 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA enzyme substrate (biology), substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each backbone chain, sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
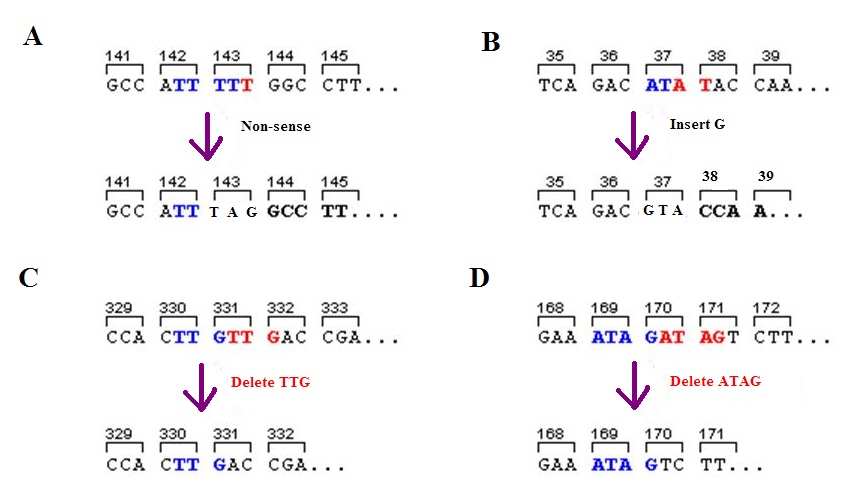
Frameshift Mutation
A frameshift mutation (also called a framing error or a reading frame shift) is a genetic mutation caused by indels ( insertions or deletions) of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet nature of gene expression by codons, the insertion or deletion can change the reading frame (the grouping of the codons), resulting in a completely different translation from the original. The earlier in the sequence the deletion or insertion occurs, the more altered the protein. A frameshift mutation is not the same as a single-nucleotide polymorphism in which a nucleotide is replaced, rather than inserted or deleted. A frameshift mutation will in general cause the reading of the codons after the mutation to code for different amino acids. The frameshift mutation will also alter the first stop codon ("UAA", "UGA" or "UAG") encountered in the sequence. The polypeptide being created could be abnormally short or abnormally long, and will most li ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-homologous End Joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. It is called "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair (HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber. NHEJ is typically guided by short homologous DNA sequences called microhomologies. These microhomologies are often present in single-stranded overhangs on the ends of double-strand breaks. When the overhangs are perfectly compatible, NHEJ usually repairs the break accurately. Imprecise repair leading to loss of nucleotides can also occur, but is much more common when the overhangs are not compatible. Inappropriate NHEJ can lead to translocations and telomere fusion, hallmarks of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Frameshift Mutations (13080927393)
A frameshift mutation (also called a framing error or a reading frame shift) is a genetic mutation caused by indels ( insertions or deletions) of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet nature of gene expression by codons, the insertion or deletion can change the reading frame (the grouping of the codons), resulting in a completely different translation from the original. The earlier in the sequence the deletion or insertion occurs, the more altered the protein. A frameshift mutation is not the same as a single-nucleotide polymorphism in which a nucleotide is replaced, rather than inserted or deleted. A frameshift mutation will in general cause the reading of the codons after the mutation to code for different amino acids. The frameshift mutation will also alter the first stop codon ("UAA", "UGA" or "UAG") encountered in the sequence. The polypeptide being created could be abnormally short or abnormally long, and will most like ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homozygous
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism. Most eukaryotes have two matching sets of chromosomes; that is, they are diploid. Diploid organisms have the same loci on each of their two sets of homologous chromosomes except that the sequences at these loci may differ between the two chromosomes in a matching pair and that a few chromosomes may be mismatched as part of a chromosomal sex-determination system. If both alleles of a diploid organism are the same, the organism is homozygous at that locus. If they are different, the organism is heterozygous at that locus. If one allele is missing, it is hemizygous, and, if both alleles are missing, it is nullizygous. The DNA sequence of a gene often varies from one individual to another. These gene variants are called alleles. While some ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Selective Breeding
Selective breeding (also called artificial selection) is the process by which humans use animal breeding and plant breeding to selectively develop particular phenotypic traits (characteristics) by choosing which typically animal or plant males and females will sexually reproduce and have offspring together. Domesticated animals are known as breeds, normally bred by a professional breeder, while domesticated plants are known as varieties, cultigens, cultivars, or breeds. Two purebred animals of different breeds produce a crossbreed, and crossbred plants are called hybrids. Flowers, vegetables and fruit-trees may be bred by amateurs and commercial or non-commercial professionals: major crops are usually the provenance of the professionals. In animal breeding artificial selection is often combined with techniques such as inbreeding, linebreeding, and outcrossing. In plant breeding, similar methods are used. Charles Darwin discussed how selective breeding had been succ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Diploid
Ploidy () is the number of complete sets of chromosomes in a cell, and hence the number of possible alleles for autosomal and pseudoautosomal genes. Here ''sets of chromosomes'' refers to the number of maternal and paternal chromosome copies, respectively, in each homologous chromosome pair—the form in which chromosomes naturally exist. Somatic cells, tissues, and individual organisms can be described according to the number of sets of chromosomes present (the "ploidy level"): monoploid (1 set), diploid (2 sets), triploid (3 sets), tetraploid (4 sets), pentaploid (5 sets), hexaploid (6 sets), heptaploid or septaploid (7 sets), etc. The generic term polyploid is often used to describe cells with three or more sets of chromosomes. Virtually all sexually reproducing organisms are made up of somatic cells that are diploid or greater, but ploidy level may vary widely between different organisms, between different tissues within the same organism, and at different stages in a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
In Vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, and plants, as opposed to a tissue extract or dead organism. Examples of investigations ''in vivo'' include: the pathogenesis of disease by comparing the effects of bacterial infection with the effects of purified bacterial toxins; the development of non-antibiotics, antiviral drugs, and new drugs generally; and new surgical procedures. Consequently, animal testing and clinical trials are major elements of ''in vivo'' research. ''In vivo'' testing is often employed over ''in vitro'' because it is better suited for observing the overall effects of an experiment on a living subject. In drug discovery, for example, verification of efficacy ''in vivo'' is crucial, because ''in vitro'' assays can sometimes yield misleading results with drug c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |