|
Fixation (population Genetics)
In population genetics, fixation is the change in a gene pool from a situation where there exists at least two variants of a particular gene (allele) in a given population to a situation where only one of the alleles remains. That is, the allele becomes fixed. In the absence of mutation or heterozygote advantage, any allele must eventually either be lost completely from the population, or fixed, i.e. permanently established at 100% frequency in the population. Whether a gene will ultimately be lost or fixed is dependent on selection coefficients and chance fluctuations in allelic proportions. Fixation can refer to a gene in general or particular nucleotide position in the DNA chain ( locus). In the process of substitution, a previously non-existent allele arises by mutation and undergoes fixation by spreading through the population by random genetic drift or positive selection. Once the frequency of the allele is at 100%, i.e. being the only gene variant present in any memb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Population Genetics
Population genetics is a subfield of genetics that deals with genetic differences within and among populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as Adaptation (biology), adaptation, speciation, and population stratification, population structure. Population genetics was a vital ingredient in the emergence of the Modern synthesis (20th century), modern evolutionary synthesis. Its primary founders were Sewall Wright, J. B. S. Haldane and Ronald Fisher, who also laid the foundations for the related discipline of quantitative genetics. Traditionally a highly mathematical discipline, modern population genetics encompasses theoretical, laboratory, and field work. Population genetic models are used both for statistical inference from DNA sequence data and for proof/disproof of concept. What sets population genetics apart from newer, more phenotypic approaches to modelling evolution, such as evolutionary game theory and evolu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Natural Selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the Heredity, heritable traits characteristic of a population over generations. Charles Darwin popularised the term "natural selection", contrasting it with selective breeding, artificial selection, which is intentional, whereas natural selection is not. Genetic diversity, Variation of traits, both Genotype, genotypic and phenotypic, exists within all populations of organisms. However, some traits are more likely to facilitate survival and reproductive success. Thus, these traits are passed the next generation. These traits can also become more Allele frequency, common within a population if the environment that favours these traits remains fixed. If new traits become more favoured due to changes in a specific Ecological niche, niche, microevolution occurs. If new traits become more favoured due to changes in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Hitchhiking
Genetic hitchhiking, also called genetic draft or the hitchhiking effect, is when an allele changes frequency not because it itself is under natural selection, but because it is near another gene that is undergoing a selective sweep and that is on the same DNA chain. When one gene goes through a selective sweep, any other nearby polymorphisms that are in linkage disequilibrium will tend to change their allele frequencies too.Futuyma, Douglas J. 2013. Evolution: Third Edition. Sinauer Associates, Inc: Sunderland, MA. Selective sweeps happen when newly appeared (and hence still rare) mutations are advantageous and increase in frequency. Neutral or even slightly deleterious alleles that happen to be close by on the chromosome 'hitchhike' along with the sweep. In contrast, effects on a neutral locus due to linkage disequilibrium with newly appeared deleterious mutations are called background selection. Both genetic hitchhiking and background selection are stochastic (random) ev ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Idealised Population
In population genetics an idealised population is one that can be described using a number of simplifying assumptions. Models of idealised populations are either used to make a general point, or they are fit to data on real populations for which the assumptions may not hold true. For example, coalescent theory is used to fit data to models of idealised populations. The most common idealized population in population genetics is described in the Wright-Fisher model after Sewall Wright and Ronald Fisher (1922, 1930) and (1931). Wright-Fisher populations have constant size, and their members can mate and reproduce with any other member. Another example is a Moran model, which has overlapping generations, rather than the non-overlapping generations of the Fisher-Wright model. The complexities of real populations can cause their behavior to match an idealised population with an effective population size that is very different from the census population size of the real population. For ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Effective Population Size
The effective population size (''N''''e'') is the size of an idealised population that would experience the same rate of genetic drift as the real population. Idealised populations are those following simple one- locus models that comply with assumptions of the neutral theory of molecular evolution. The effective population size is normally smaller than the census population size ''N'', partly because chance events prevent some individuals from breeding, and partly due to background selection and genetic hitchhiking. The same real population could have a different effective population size for different properties of interest, such as genetic drift (or more precisely, the speed of coalescence) over one generation vs. over many generations. Within a species, areas of the genome that have more genes and/or less genetic recombination tend to have lower effective population sizes, because of the effects of selection at linked sites. In a population with selection at many loci an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
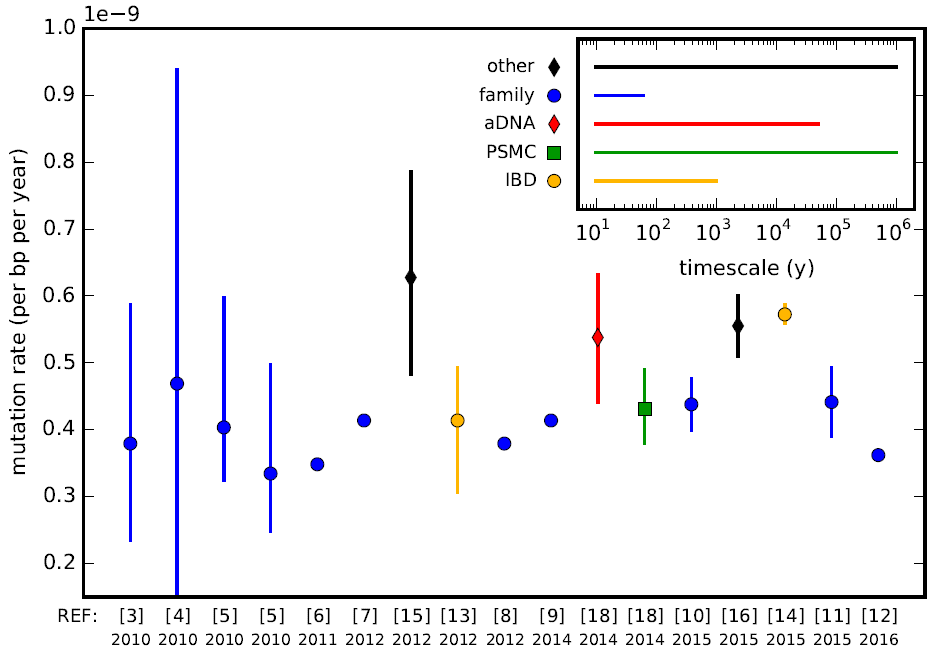
Mutation Rate
In genetics, the mutation rate is the frequency of new mutations in a single gene, nucleotide sequence, or organism over time. Mutation rates are not constant and are not limited to a single type of mutation; there are many different types of mutations. Mutation rates are given for specific classes of mutations. Point mutations are a class of mutations that are changes to a single base. Missense, nonsense, and synonymous mutations are three subtypes of point mutations. The rate of these types of substitutions can be further subdivided into a mutation spectrum, which describes the influence of the genetic context on the mutation rate. There are several natural units of time for each of these rates, with rates being characterized either as mutations per base pair per cell division, per gene per generation, or genome per generation. The mutation rate of an organism is an evolved characteristic and is strongly influenced by the genetics of each organism, in addition to a strong in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Diploid
Ploidy () is the number of complete sets of chromosomes in a cell, and hence the number of possible alleles for autosomal and pseudoautosomal genes. Here ''sets of chromosomes'' refers to the number of maternal and paternal chromosome copies, respectively, in each homologous chromosome pair—the form in which chromosomes naturally exist. Somatic cells, tissues, and individual organisms can be described according to the number of sets of chromosomes present (the "ploidy level"): monoploid (1 set), diploid (2 sets), triploid (3 sets), tetraploid (4 sets), pentaploid (5 sets), hexaploid (6 sets), heptaploid or septaploid (7 sets), etc. The generic term polyploid is often used to describe cells with three or more sets of chromosomes. Virtually all sexually reproducing organisms are made up of somatic cells that are diploid or greater, but ploidy level may vary widely between different organisms, between different tissues within the same organism, and at different stages in a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Motoo Kimura
(November 13, 1924 – November 13, 1994) was a Japanese biologist best known for introducing the neutral theory of molecular evolution in 1968. He became one of the most influential theoretical population geneticists. He is remembered in genetics for his innovative use of diffusion equations to calculate the probability of fixation of beneficial, deleterious, or neutral alleles. Combining theoretical population genetics with molecular evolution data, he also developed the neutral theory of molecular evolution in which genetic drift is the main force changing allele frequencies. James F. Crow, himself a renowned population geneticist, considered Kimura to be one of the two greatest evolutionary geneticists, along with Gustave Malécot, after the great trio of the modern synthesis, Ronald Fisher, J. B. S. Haldane, and Sewall Wright. Life and work Kimura was born on November 13, 1924, in Okazaki, Aichi Prefecture. From an early age he was very interested in botany, thoug ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Species
A species () is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. It is the basic unit of Taxonomy (biology), classification and a taxonomic rank of an organism, as well as a unit of biodiversity. Other ways of defining species include their karyotype, DNA sequence, morphology (biology), morphology, behaviour, or ecological niche. In addition, palaeontologists use the concept of the chronospecies since fossil reproduction cannot be examined. The most recent rigorous estimate for the total number of species of eukaryotes is between 8 and 8.7 million. About 14% of these had been described by 2011. All species (except viruses) are given a binomial nomenclature, two-part name, a "binomen". The first part of a binomen is the name of a genus to which the species belongs. The second part is called the specific name (zoology), specific name or the specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Taxa
In biology, a taxon (back-formation from ''taxonomy''; : taxa) is a group of one or more populations of an organism or organisms seen by taxonomists to form a unit. Although neither is required, a taxon is usually known by a particular name and given a particular ranking, especially if and when it is accepted or becomes established. It is very common, however, for taxonomists to remain at odds over what belongs to a taxon and the criteria used for inclusion, especially in the context of rank-based (" Linnaean") nomenclature (much less so under phylogenetic nomenclature). If a taxon is given a formal scientific name, its use is then governed by one of the nomenclature codes specifying which scientific name is correct for a particular grouping. Initial attempts at classifying and ordering organisms (plants and animals) were presumably set forth in prehistoric times by hunter-gatherers, as suggested by the fairly sophisticated folk taxonomies. Much later, Aristotle, and later st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allele Frequency
Allele frequency, or gene frequency, is the relative frequency of an allele (variant of a gene) at a particular locus in a population, expressed as a fraction or percentage. Specifically, it is the fraction of all chromosomes in the population that carry that allele over the total population or sample size. Microevolution is the change in allele frequencies that occurs over time within a population. Given the following: # A particular locus on a chromosome and a given allele at that locus # A population of ''N'' individuals with ploidy ''n'', i.e. an individual carries ''n'' copies of each chromosome in their somatic cells (e.g. two chromosomes in the cells of diploid species) # The allele exists in ''i'' chromosomes in the population then the allele frequency is the fraction of all the occurrences ''i'' of that allele and the total number of chromosome copies across the population, ''i''/(''nN''). The allele frequency is distinct from the genotype frequency, although they a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance. Genetic drift may cause gene variants to disappear completely and thereby reduce genetic variation. It can also cause initially rare alleles to become much more frequent and even fixed. When few copies of an allele exist, the effect of genetic drift is more notable, and when many copies exist, the effect is less notable (due to the law of large numbers). In the middle of the 20th century, vigorous debates occurred over the relative importance of natural selection versus neutral processes, including genetic drift. Ronald Fisher, who explained natural selection using Mendelian inheritance, Mendelian genetics, held the view that genetic drift plays at most a minor role in evolution, and this remained the dominant view for several decades. In 1968, population geneticist Mot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |