|
Fine Structure Genetics
{{One source, date=December 2016 Fine structure genetics is a subfield of genetics which encompasses a set of tools used to examine not just the mutations within an entire genome, but can be isolated to either specific pathways or regions of the genome. Ultimately, this more focused lens can lead to a more nuanced and interactive view of the function of a gene. Regional Mutagenesis Similar to forward genetics, regional mutagenesis seeks to saturate with insertions or point mutations, but instead of for the entire genome, it saturates only a small portion of the genome. By limiting the region in focus, researchers are then able to intensify the number of mutations within any genes or promoters within that regions, often illuminating more complicated functions than could be identified with a broader focus. Furthermore, such mutations can show how the specific structure of that region of a chromosome affect expression levels and function.A Primer of Genome Science, Third Edition. Gr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinians, Augustinian friar working in the 19th century in Brno, was the first to study genetics scientifically. Mendel studied "trait inheritance", patterns in the way traits are handed down from parents to offspring over time. He observed that organisms (pea plants) inherit traits by way of discrete "units of inheritance". This term, still used today, is a somewhat ambiguous definition of what is referred to as a gene. Phenotypic trait, Trait inheritance and Molecular genetics, molecular inheritance mechanisms of genes are still primary principles of genetics in the 21st century, but modern genetics has expanded to study the function and behavior of genes. Gene structure and function, variation, and distribution are studied within the context of the Cell (bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA (such as pyrimidine dimers caused by exposure to ultraviolet radiation), which then may undergo error-prone repair (especially microhomology-mediated end joining), cause an error during other forms of repair, or cause an error during replication ( translesion synthesis). Mutations may also result from substitution, insertion or deletion of segments of DNA due to mobile genetic elements. Mutations may or may not produce detectable changes in the observable characteristics ( phenotype) of an organism. Mutations play a part in both normal and abnormal biological processes including: evolution, cancer, and the development of the immune system, including junctional diversity. Mutati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondrial DNA, mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplast DNA, chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been Whole-genome sequencing, sequenced and various regions have been annotated. The first genome to be sequenced was that of the virus φX174 in 1977; the first genome sequence of a prokaryote (''Haemophilus influenzae'') was published in 1995; the yeast (''Saccharomyces cerevisiae'') genome was the first eukaryotic genome to be sequenced in 1996. The Human Genome Project ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Regulatory Network
A gene (or genetic) regulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the function of the cell. GRN also play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo). The regulator can be DNA, RNA, protein or any combination of two or more of these three that form a complex, such as a specific sequence of DNA and a transcription factor to activate that sequence. The interaction can be direct or indirect (through transcribed RNA or translated protein). In general, each mRNA molecule goes on to make a specific protein (or set of proteins). In some cases this protein will be structural, and will accumulate at the cell membrane or within the cell to give it particular structural properties. In other cases the protein will be an enz ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes. During gene expression (the synthesis of Gene product, RNA or protein from a gene), DNA is first transcription (biology), copied into RNA. RNA can be non-coding RNA, directly functional or be the intermediate protein biosynthesis, template for the synthesis of a protein. The transmission of genes to an organism's offspring, is the basis of the inheritance of phenotypic traits from one generation to the next. These genes make up different DNA sequences, together called a genotype, that is specific to every given individual, within the gene pool of the population (biology), population of a given species. The genotype, along with environmental and developmental factors, ultimately determines the phenotype ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Forward Genetics
Forward genetics is a molecular genetics approach of determining the genetic basis responsible for a phenotype. Forward genetics provides an unbiased approach because it relies heavily on identifying the genes or genetic factors that cause a particular phenotype or trait of interest. This was initially done by using naturally occurring mutations or inducing mutants with radiation, chemicals, or insertional mutagenesis (e.g. transposable elements). Subsequent breeding takes place, mutant individuals are isolated, and then the gene is gene mapping, mapped. Forward genetics can be thought of as a counter to reverse genetics, which determines the function of a gene by analyzing the phenotypic effects of altered DNA sequences. Mutant phenotypes are often observed long before having any idea which gene is responsible, which can lead to genes being named after their mutant phenotype (e.g. Drosophila ''rosy'' gene which is named after the eye colour in mutants). Techniques used in Forward ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transposable Element
A transposable element (TE), also transposon, or jumping gene, is a type of mobile genetic element, a nucleic acid sequence in DNA that can change its position within a genome. The discovery of mobile genetic elements earned Barbara McClintock a Nobel Prize in 1983. There are at least two classes of TEs: Class I TEs or retrotransposons generally function via reverse transcription, while Class II TEs or DNA transposons encode the protein transposase, which they require for insertion and excision, and some of these TEs also encode other proteins. Discovery by Barbara McClintock Barbara McClintock discovered the first TEs in maize (''Zea mays'') at the Cold Spring Harbor Laboratory in New York. McClintock was experimenting with maize plants that had broken chromosomes. In the winter of 1944–1945, McClintock planted corn kernels that were self-pollinated, meaning that the silk (style) of the flower received pollen from its own anther. These kernels came from a long line ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Balancer Chromosomes
Balancer chromosomes (or simply balancers) are a type of genetic engineering, genetically engineered chromosome used in laboratory biology for the maintenance of Lethal allele#Recessive lethals, recessive lethal (or sterile) mutations within living organisms without interference from natural selection. Since such mutations are viable only in heterozygotes, they cannot be stably maintained through successive generations and therefore continually lead to the production of wild-type organisms, which can be prevented by replacing the homologous chromosome, homologous wild-type chromosome with a balancer. In this capacity, balancers are crucial for genetics research on model organisms such as ''Drosophila melanogaster'', the common fruit fly, for which stocks cannot be archived (e.g. frozen). They can also be used in forward genetics genetic screening, screens to specifically identify recessive lethal (or sterile) mutations. For that reason, balancers are also used in other model organism ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enhancer Trap
An enhancer trap is a method in molecular biology. The enhancer trap construct contains a transposable element and a reporter gene. The first is necessary for (random) insertion in the genome, the latter is necessary for identification of the spatial regulation by the enhancer. On top of this, the construct usually includes a genetic marker, e.g., the White (mutation), white gene producing red-colored eyes in ''Drosophila'', or ampicillin resistance in ''Escherichia coli, E. coli''. The most common and basic enhancer traps are: P[lacZ] from the bacterium ''Escherichia coli, E. coli'' and P[GAL4/UAS system, GAL4] from yeast. There exists a large number of fly stocks containing GAL4 insertions and an equally large number of fly stocks containing an UAS DNA sequence followed by a gene of interest, which permits the expression of a large number of genes with different GAL4 "drivers". Rather than generating transgenic flies with the enhancer linked directly to the gene of interest (which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Green Fluorescent Protein
The green fluorescent protein (GFP) is a protein that exhibits green fluorescence when exposed to light in the blue to ultraviolet range. The label ''GFP'' traditionally refers to the protein first isolated from the jellyfish ''Aequorea victoria'' and is sometimes called ''avGFP''. However, GFPs have been found in other organisms including corals, sea anemones, zoanithids, copepods and lancelets. The GFP from ''A. victoria'' has a major excitation peak at a wavelength of 395 nm and a minor one at 475 nm. Its emission peak is at 509 nm, which is in the lower green portion of the visible spectrum. The fluorescence quantum yield (QY) of GFP is 0.79. The GFP from the sea pansy ('' Renilla reniformis'') has a single major excitation peak at 498 nm. GFP makes for an excellent tool in many forms of biology due to its ability to form an internal chromophore without requiring any accessory cofactors, gene products, or enzymes / substrates other than molecular ox ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
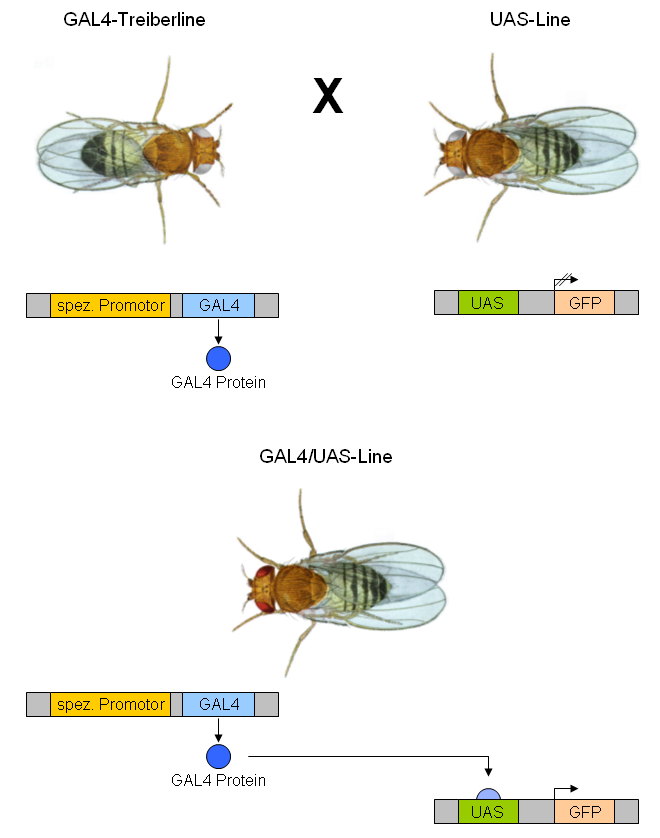
GAL4/UAS System
The GAL4-UAS system is a biochemical method used to study gene expression and function in organisms such as the Drosophila melanogaster, fruit fly. It is based on the finding by Hitoshi Kakidani and Mark Ptashne, and Nicholas Webster and Pierre Chambon in 1988 that Gal4 binding to UAS sequences activates gene expression. The method was introduced into flies by Andrea Brand and Norbert Perrimon in 1993 and is considered a powerful technique for studying the Gene expression, expression of genes. The system has two parts: the Gal4 transcription factor, Gal4 gene, encoding the Saccharomyces cerevisiae, yeast transcription activator protein Gal4 transcription factor, Gal4, and the UAS (Upstream Activation Sequence), an enhancer to which GAL4 specifically binds to activate gene transcription (genetics), transcription. Overview The Gal4 transcription factor, Gal4 system allows separation of the problems of defining which cells express a gene or protein and what the experimenter wants to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |