|
Expression Profiling
In the field of molecular biology, gene expression profiling is the measurement of the activity (the expression) of thousands of genes at once, to create a global picture of cellular function. These profiles can, for example, distinguish between cells that are actively dividing, or show how the cells react to a particular treatment. Many experiments of this sort measure an entire genome simultaneously, that is, every gene present in a particular cell. Several transcriptomics technologies can be used to generate the necessary data to analyse. DNA microarrays measure the relative activity of previously identified target genes. Sequence based techniques, like RNA-Seq, provide information on the sequences of genes in addition to their expression level. Background Expression profiling is a logical next step after sequencing a genome: the sequence tells us what the cell could possibly do, while the expression profile tells us what it is actually doing at a point in time. Genes conta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heatmap
A heat map (or heatmap) is a data visualization technique that shows magnitude of a phenomenon as color in two dimensions. The variation in color may be by hue or brightness, intensity, giving obvious visual cues to the reader about how the phenomenon is clustered or varies over space. There are two fundamentally different categories of heat maps: the cluster heat map and the spatial heat map. In a cluster heat map, magnitudes are laid out into a matrix of fixed cell size whose rows and columns are discrete phenomena and categories, and the sorting of rows and columns is intentional and somewhat arbitrary, with the goal of suggesting clusters or portraying them as discovered via statistical analysis. The size of the cell is arbitrary but large enough to be clearly visible. By contrast, the position of a magnitude in a spatial heat map is forced by the location of the magnitude in that space, and there is no notion of cells; the phenomenon is considered to vary continuously. "Heat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Posttranslational Modification
Post-translational modification (PTM) is the covalent and generally enzymatic modification of proteins following protein biosynthesis. This process occurs in the endoplasmic reticulum and the golgi apparatus. Proteins are synthesized by ribosomes translating mRNA into polypeptide chains, which may then undergo PTM to form the mature protein product. PTMs are important components in cell signaling, as for example when prohormones are converted to hormones. Post-translational modifications can occur on the amino acid side chains or at the protein's C- or N- termini. They can extend the chemical repertoire of the 20 standard amino acids by modifying an existing functional group or introducing a new one such as phosphate. Phosphorylation is a highly effective mechanism for regulating the activity of enzymes and is the most common post-translational modification. Many eukaryotic and prokaryotic proteins also have carbohydrate molecules attached to them in a process called gl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
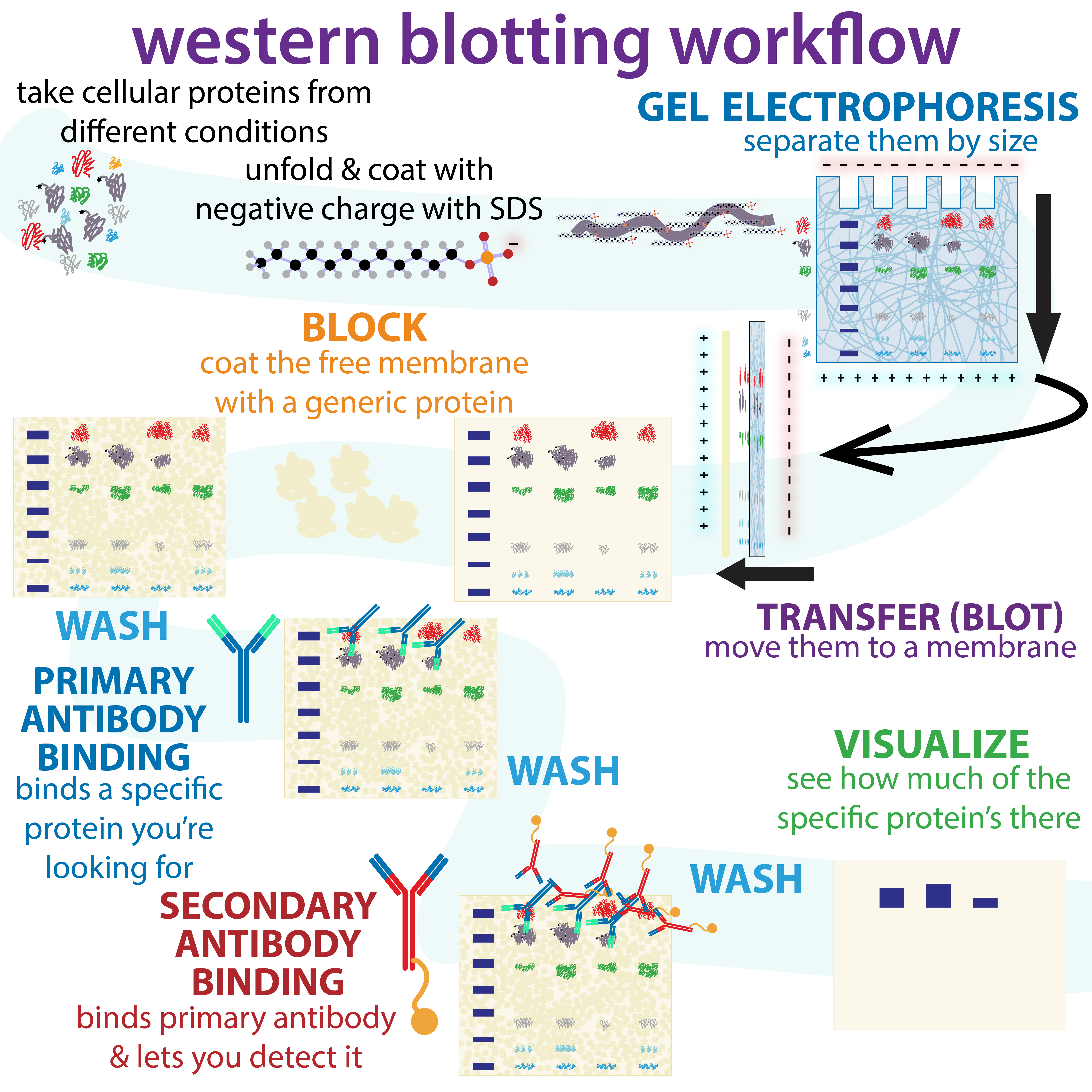
Western Blot
The western blot (sometimes called the protein immunoblot), or western blotting, is a widely used analytical technique in molecular biology and immunogenetics to detect specific proteins in a sample of tissue homogenate or extract. Besides detecting the proteins, this technique is also utilized to visualize, distinguish, and quantify the different proteins in a complicated protein combination. Western blot technique uses three elements to achieve its task of separating a specific protein from a complex: separation by size, transfer of protein to a solid support, and marking target protein using a primary and secondary antibody to visualize. A synthetic or animal-derived antibody (known as the primary antibody) is created that recognizes and binds to a specific target protein. The electrophoresis membrane is washed in a solution containing the primary antibody, before excess antibody is washed off. A secondary antibody is added which recognizes and binds to the primary antibod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Base Pairing
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine–thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
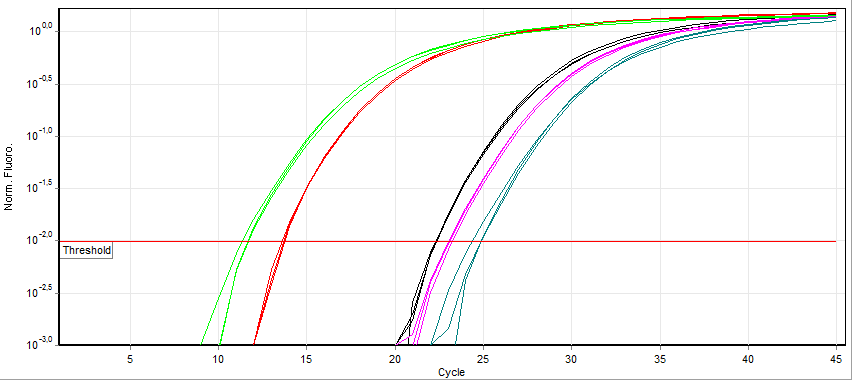
Quantitative PCR
A real-time polymerase chain reaction (real-time PCR, or qPCR) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively (quantitative real-time PCR) and semi-quantitatively (i.e., above/below a certain amount of DNA molecules) (semi-quantitative real-time PCR). Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter, which permits detection only after hybridization of the probe with its complementary sequence. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines propose that the abbreviation ''qPCR'' be used for qu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microarray Databases
A microarray database is a repository containing microarray gene expression data. The key uses of a microarray database are to store the measurement data, manage a searchable index, and make the data available to other applications for analysis and interpretation (either directly, or via user downloads). Microarray databases can fall into two distinct classes: # A peer reviewed, public repository that adheres to academic or industry standards and is designed to be used by many analysis applications and groups. A good example of this is the Gene Expression Omnibus (GEO) from NCBI or ArrayExpress from EBI. # A specialized repository associated primarily with the brand of a particular entity (lab, company, university, consortium, group), an application suite, a topic, or an analysis method, whether it is commercial, non-profit, or academic. These databases might have one or more of the following characteristics: #* A subscription or license may be needed to gain full access, #* The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microarray Analysis Techniques
Microarray analysis techniques are used in interpreting the data generated from experiments on DNA (Gene chip analysis), RNA, and protein microarrays, which allow researchers to investigate the expression state of a large number of genes - in many cases, an organism's entire genome - in a single experiment. Such experiments can generate very large amounts of data, allowing researchers to assess the overall state of a cell or organism. Data in such large quantities is difficult - if not impossible - to analyze without the help of computer programs. Introduction Microarray data analysis is the final step in reading and processing data produced by a microarray chip. Samples undergo various processes including purification and scanning using the microchip, which then produces a large amount of data that requires processing via computer software. It involves several distinct steps, as outlined in the image below. Changing any one of the steps will change the outcome of the analysis, so ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
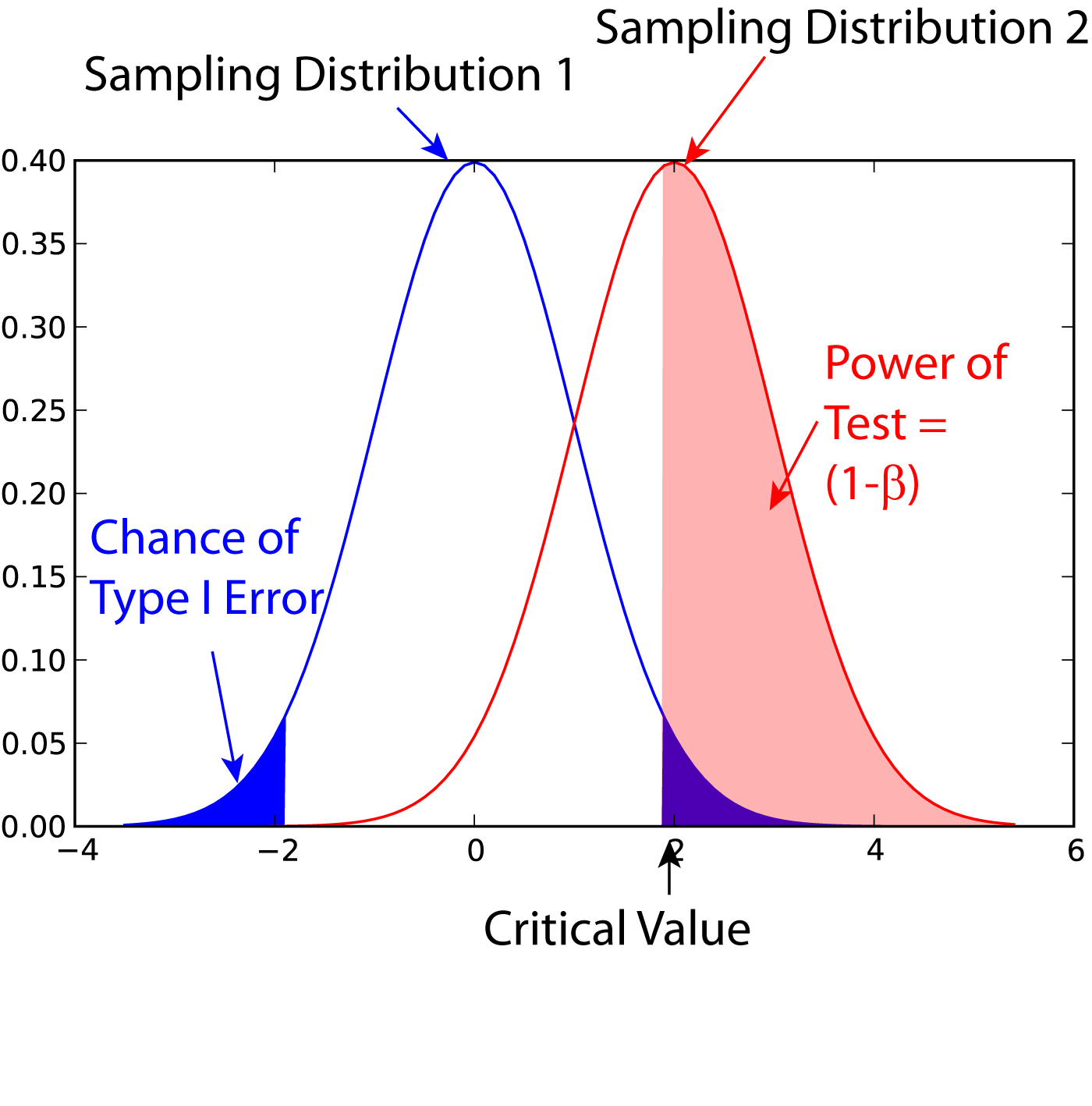
Statistical Power
In statistics, the power of a binary hypothesis test is the probability that the test correctly rejects the null hypothesis (H_0) when a specific alternative hypothesis (H_1) is true. It is commonly denoted by 1-\beta, and represents the chances of a true positive detection conditional on the actual existence of an effect to detect. Statistical power ranges from 0 to 1, and as the power of a test increases, the probability \beta of making a type II error by wrongly failing to reject the null hypothesis decreases. Notation This article uses the following notation: * ''β'' = probability of a Type II error, known as a "false negative" * 1 − ''β'' = probability of a "true positive", i.e., correctly rejecting the null hypothesis. "1 − ''β''" is also known as the power of the test. * ''α'' = probability of a Type I error, known as a "false positive" * 1 − ''α'' = probability of a "true negative", i.e., correctly not rejecting the null hypothesis Description For a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cellular Differentiation
Cellular differentiation is the process in which a stem cell alters from one type to a differentiated one. Usually, the cell changes to a more specialized type. Differentiation happens multiple times during the development of a multicellular organism as it changes from a simple zygote to a complex system of tissues and cell types. Differentiation continues in adulthood as adult stem cells divide and create fully differentiated daughter cells during tissue repair and during normal cell turnover. Some differentiation occurs in response to antigen exposure. Differentiation dramatically changes a cell's size, shape, membrane potential, metabolic activity, and responsiveness to signals. These changes are largely due to highly controlled modifications in gene expression and are the study of epigenetics. With a few exceptions, cellular differentiation almost never involves a change in the DNA sequence itself. Although metabolic composition does get altered quite dramatical ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cross-validation (statistics)
Cross-validation, sometimes called rotation estimation or out-of-sample testing, is any of various similar model validation techniques for assessing how the results of a statistical analysis will generalize to an independent data set. Cross-validation is a resampling method that uses different portions of the data to test and train a model on different iterations. It is mainly used in settings where the goal is prediction, and one wants to estimate how accurately a predictive model will perform in practice. In a prediction problem, a model is usually given a dataset of ''known data'' on which training is run (''training dataset''), and a dataset of ''unknown data'' (or ''first seen'' data) against which the model is tested (called the validation dataset or ''testing set''). The goal of cross-validation is to test the model's ability to predict new data that was not used in estimating it, in order to flag problems like overfitting or selection bias and to give an insight on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Markov Clustering
In statistics, Markov chain Monte Carlo (MCMC) methods comprise a class of algorithms for sampling from a probability distribution. By constructing a Markov chain that has the desired distribution as its equilibrium distribution, one can obtain a sample of the desired distribution by recording states from the chain. The more steps that are included, the more closely the distribution of the sample matches the actual desired distribution. Various algorithms exist for constructing chains, including the Metropolis–Hastings algorithm. Application domains MCMC methods are primarily used for calculating numerical approximations of multi-dimensional integrals, for example in Bayesian statistics, computational physics, computational biology and computational linguistics. In Bayesian statistics, the recent development of MCMC methods has made it possible to compute large hierarchical models that require integrations over hundreds to thousands of unknown parameters. In rare e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hierarchical Clustering
In data mining and statistics, hierarchical clustering (also called hierarchical cluster analysis or HCA) is a method of cluster analysis that seeks to build a hierarchy of clusters. Strategies for hierarchical clustering generally fall into two categories: * Agglomerative: This is a " bottom-up" approach: Each observation starts in its own cluster, and pairs of clusters are merged as one moves up the hierarchy. * Divisive: This is a "top-down" approach: All observations start in one cluster, and splits are performed recursively as one moves down the hierarchy. In general, the merges and splits are determined in a greedy manner. The results of hierarchical clustering are usually presented in a dendrogram. The standard algorithm for hierarchical agglomerative clustering (HAC) has a time complexity of \mathcal(n^3) and requires \Omega(n^2) memory, which makes it too slow for even medium data sets. However, for some special cases, optimal efficient agglomerative methods (of comp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |