|
Docking@Home
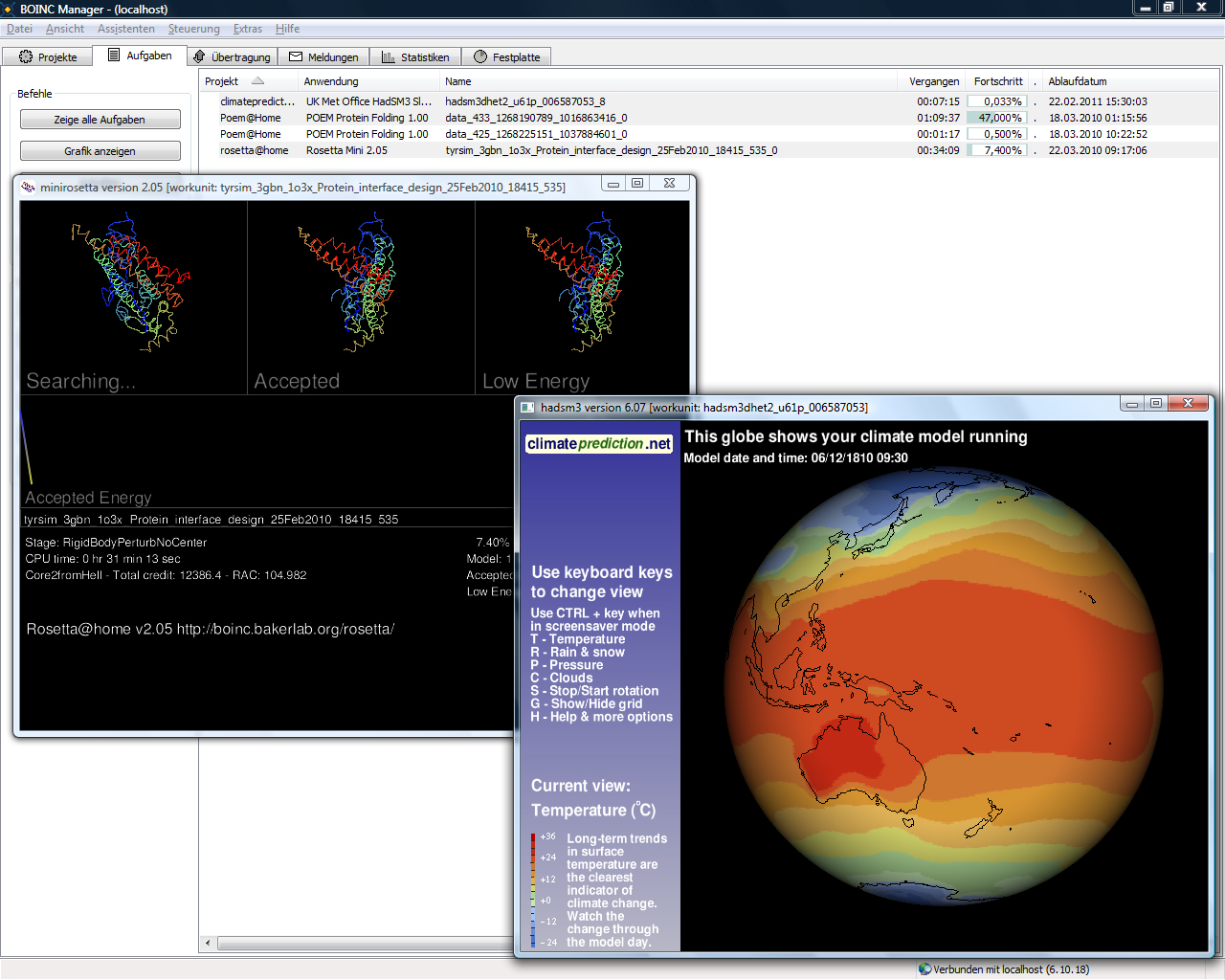
Docking@Home was a volunteer computing project hosted by the University of Delaware and running on the Berkeley Open Infrastructure for Network Computing (BOINC) software platform. It models protein-ligand docking using the CHARMM program. Volunteer computing allows an extensive search of protein-ligand docking conformations and selection of near-native ligand conformations are achieved by using ligand based hierarchical clustering. The ultimate aim was the development of new pharmaceutical drugs. The project was retired on May 23, 2014. See also *List of volunteer computing projects This is a comprehensive list of volunteer computing projects; a type of distributed computing where volunteers donate computing time to specific causes. The donated computing power comes from idle CPUs and GPUs in personal computers, video gam ... References Further reading * External links * * Science in society Volunteer computing projects Internet properties dise ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Volunteer Computing Projects
This is a comprehensive list of volunteer computing projects; a type of distributed computing where volunteers donate computing time to specific causes. The donated computing power comes from idle CPUs and GPUs in personal computers, video game consoles and Android devices. Each project seeks to utilize the computing power of many internet connected devices to solve problems and perform tedious, repetitive research in a very cost effective manner. Active projects Completed projects See also * List of grid computing projects * List of citizen science projects * List of crowdsourcing projects * List of free and open-source Android applications This is a list of notable applications (''apps'') that run on the Android platform which meet guidelines for free software and open-source software. Advertisement blocking Web browsers Office Suites and synchronisation C ... * List of Berkeley Open Infrastructure for Network Computing (BOINC) pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BOINC
The Berkeley Open Infrastructure for Network Computing (BOINC, pronounced – rhymes with "oink") is an open-source middleware system for volunteer computing (a type of distributed computing). Developed originally to support SETI@home, it became the platform for many other applications in areas as diverse as medicine, molecular biology, mathematics, linguistics, climatology, environmental science, and astrophysics, among others. The purpose of BOINC is to enable researchers to utilize processing resources of personal computers and other devices around the world. BOINC development began with a group based at the Space Sciences Laboratory (SSL) at the University of California, Berkeley, and led by David P. Anderson, who also led SETI@home. As a high-performance volunteer computing platform, BOINC brings together 34,236 active participants employing 136,341 active computers (hosts) worldwide, processing daily on average 20.164 PetaFLOPS (it would be the 21st largest processin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Berkeley Open Infrastructure For Network Computing
The Berkeley Open Infrastructure for Network Computing (BOINC, pronounced – rhymes with "oink") is an open-source middleware system for volunteer computing (a type of distributed computing). Developed originally to support SETI@home, it became the platform for many other applications in areas as diverse as medicine, molecular biology, mathematics, linguistics, climatology, environmental science, and astrophysics, among others. The purpose of BOINC is to enable researchers to utilize processing resources of personal computers and other devices around the world. BOINC development began with a group based at the Space Sciences Laboratory (SSL) at the University of California, Berkeley, and led by David P. Anderson, who also led SETI@home. As a high-performance volunteer computing platform, BOINC brings together 34,236 active participants employing 136,341 active computers (hosts) worldwide, processing daily on average 20.164 PetaFLOPS (it would be the 21st largest processin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CHARMM
Chemistry at Harvard Macromolecular Mechanics (CHARMM) is the name of a widely used set of force fields for molecular dynamics, and the name for the molecular dynamics simulation and analysis computer software package associated with them. The CHARMM Development Project involves a worldwide network of developers working with Martin Karplus and his group at Harvard to develop and maintain the CHARMM program. Licenses for this software are available, for a fee, to people and groups working in academia. Force fields The CHARMM force fields for proteins include: united-atom (sometimes termed ''extended atom'') CHARMM19, all-atom CHARMM22 and its dihedral potential corrected variant CHARMM22/CMAP, as well as later versions CHARMM27 and CHARMM36 and various modifications such as CHARMM36m and CHARMM36IDPSFF. In the CHARMM22 protein force field, the atomic partial charges were derived from quantum chemical calculations of the interactions between model compounds and water. Furthermore ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligand (biochemistry)
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure. Binding occurs by intermolecular forces, such as ionic bonds, hydrogen bonds and Van der Waals forces. The association or docking is actually reversible through dissociation. Measurably irreversible covalent bonding between a ligand and target molecule is atypical in biological systems. In contrast to the definition ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Docking (molecular)
In the field of molecular modeling, docking is a method which predicts the preferred orientation of one molecule to a second when a ligand and a target are bound to each other to form a stable complex. Knowledge of the preferred orientation in turn may be used to predict the strength of association or binding affinity between two molecules using, for example, scoring functions. The associations between biologically relevant molecules such as proteins, peptides, nucleic acids, carbohydrates, and lipids play a central role in signal transduction. Furthermore, the relative orientation of the two interacting partners may affect the type of signal produced (e.g., agonism vs antagonism). Therefore, docking is useful for predicting both the strength and type of signal produced. Molecular docking is one of the most frequently used methods in structure-based drug design, due to its ability to predict the binding-conformation of small molecule ligands to the appropriate target b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cross-platform
In computing, cross-platform software (also called multi-platform software, platform-agnostic software, or platform-independent software) is computer software that is designed to work in several computing platforms. Some cross-platform software requires a separate build for each platform, but some can be directly run on any platform without special preparation, being written in an interpreted language or compiled to portable bytecode for which the interpreters or run-time packages are common or standard components of all supported platforms. For example, a cross-platform application may run on Microsoft Windows, Linux, and macOS. Cross-platform software may run on many platforms, or as few as two. Some frameworks for cross-platform development are Codename One, Kivy, Qt, Flutter, NativeScript, Xamarin, Phonegap, Ionic, and React Native. Platforms ''Platform'' can refer to the type of processor (CPU) or other hardware on which an operating system (OS) or application runs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Volunteer Computing
Volunteer computing is a type of distributed computing in which people donate their computers' unused resources to a research-oriented project, and sometimes in exchange for credit points. The fundamental idea behind it is that a modern desktop computer is sufficiently powerful to perform billions of operations a second, but for most users only between 10-15% of its capacity is used. Typical uses like basic word processing or web browsing leave the computer mostly idle. The practice of volunteer computing, which dates back to the mid-1990s, can potentially make substantial processing power available to researchers at minimal cost. Typically, a program running on a volunteer's computer periodically contacts a research application to request jobs and report results. A middleware system usually serves as an intermediary. History The first volunteer computing project was the Great Internet Mersenne Prime Search, which was started in January 1996. It was followed in 1997 by distribute ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
University Of Delaware
The University of Delaware (colloquially UD or Delaware) is a public land-grant research university located in Newark, Delaware. UD is the largest university in Delaware. It offers three associate's programs, 148 bachelor's programs, 121 master's programs (with 13 joint degrees), and 55 doctoral programs across its eight colleges. The main campus is in Newark, with satellite campuses in Dover, Wilmington, Lewes, and Georgetown. It is considered a large institution with approximately 18,200 undergraduate and 4,200 graduate students. It is a privately governed university which receives public funding for being a land-grant, sea-grant, and space-grant state-supported research institution. UDel is ranked among the top 150 universities in the U.S. UD is classified among "R1: Doctoral Universities – Very high research activity". According to the National Science Foundation, UD spent $186 million on research and development in 2018, ranking it 119th in the nation. It is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pharmaceutical Drug
A medication (also called medicament, medicine, pharmaceutical drug, medicinal drug or simply drug) is a drug used to diagnose, cure, treat, or prevent disease. Drug therapy ( pharmacotherapy) is an important part of the medical field and relies on the science of pharmacology for continual advancement and on pharmacy for appropriate management. Drugs are classified in multiple ways. One of the key divisions is by level of control, which distinguishes prescription drugs (those that a pharmacist dispenses only on the order of a physician, physician assistant, or qualified nurse) from over-the-counter drugs (those that consumers can order for themselves). Another key distinction is between traditional small molecule drugs, usually derived from chemical synthesis, and biopharmaceuticals, which include recombinant proteins, vaccines, blood products used therapeutically (such as IVIG), gene therapy, monoclonal antibodies and cell therapy (for instance, stem cell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Science In Society
Science is a systematic endeavor that builds and organizes knowledge in the form of testable explanations and predictions about the universe. Science may be as old as the human species, and some of the earliest archeological evidence for scientific reasoning is tens of thousands of years old. The earliest written records in the history of science come from Ancient Egypt and Mesopotamia in around 3000 to 1200 BCE. Their contributions to mathematics, astronomy, and medicine entered and shaped Greek natural philosophy of classical antiquity, whereby formal attempts were made to provide explanations of events in the physical world based on natural causes. After the fall of the Western Roman Empire, knowledge of Greek conceptions of the world deteriorated in Western Europe during the early centuries (400 to 1000 CE) of the Middle Ages, but was preserved in the Muslim world during the Islamic Golden Age and later by the efforts of Byzantine Greek scholars who brought Greek man ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |