|
DeCODE Genetics
deCODE genetics () is a biopharmaceutical company based in Reykjavík, Iceland. The company was founded in 1996 by Kári Stefánsson with the aim of using population genetics studies to identify variations in the human genome associated with common diseases, and to apply these discoveries "to develop novel methods to identify, treat and prevent diseases." As of 2019, more than two-thirds of the adult population of Iceland was participating in the company's research efforts, and this "population approach" serves as a model for large-scale precision medicine and national genome projects around the world. deCODE is probably best known for its discoveries in human genetics, published in major scientific journals and widely reported in the international media. But it has also made pioneering contributions to the realization of precision medicine more broadly, through public engagement in large-scale scientific research; the development of DNA-based disease risk testing for individua ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DecodeME
DecodeME is an ongoing genome-wide association study searching for genetic risk factors for Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). With a planned recruitment of 25,000 patients, it is expected to be the largest such study to date. Recruitment closed on 15 November 2023 and results are expected in 2025. Background ME/CFS is a chronic medical condition that often causes significant disability, and whose cause is unknown. Genetic studies of ME/CFS have been done before, but without significant findings. The authors of a 2022 study suggested that research with more participants is needed to discover Statistical significance, statistically significant differences. DecodeME aims to perform such a large study. It is being run as a partnership between Action for ME and the University of Edinburgh's MRC Human Genetics Unit, with Chris Ponting as chief investigator, and with £3.2 million in funding from the UK's Medical Research Council (United Kingdom), Medica ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microsatellite
A microsatellite is a tract of repetitive DNA in which certain Sequence motif, DNA motifs (ranging in length from one to six or more base pairs) are repeated, typically 5–50 times. Microsatellites occur at thousands of locations within an organism's genome. They have a higher mutation rate than other areas of DNA leading to high genetic diversity. Microsatellites are often referred to as short tandem repeats (STRs) by forensic genetics, forensic geneticists and in genetic genealogy, or as simple sequence repeats (SSRs) by plant geneticists. Microsatellites and their longer cousins, the minisatellites, together are classified as variable number tandem repeat, VNTR (variable number of tandem repeats) DNA. The name Satellite DNA, "satellite" DNA refers to the early observation that centrifugation of genomic DNA in a test tube separates a prominent layer of bulk DNA from accompanying "satellite" layers of repetitive DNA. They are widely used for DNA profiling in Loss of heterozygos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
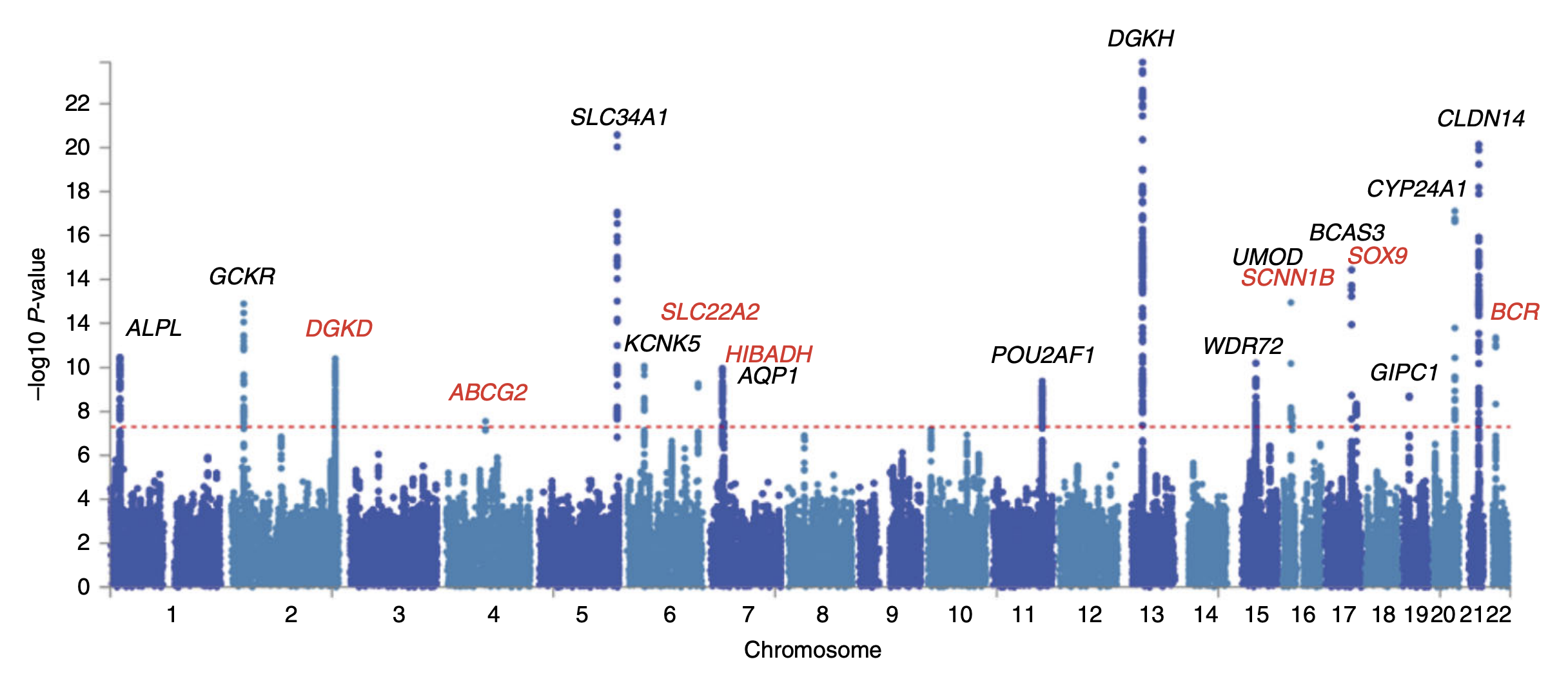
Genome-wide Association Study
In genomics, a genome-wide association study (GWA study, or GWAS), is an observational study of a genome-wide set of Single-nucleotide polymorphism, genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms. When applied to human data, GWA studies compare the DNA of participants having varying phenotypes for a particular trait or disease. These participants may be people with a disease (cases) and similar people without the disease (controls), or they may be people with different phenotypes for a particular trait, for example blood pressure. This approach is known as phenotype-first, in which the participants are classified first by their clinical manifestation(s), as opposed to Genotype-first approach, genotype-first. Each person gives a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nature Genetics
''Nature Genetics'' is a peer-reviewed scientific journal published by Nature Portfolio. It was established in 1992. It covers research in genetics. The chief editor is Tiago Faial. The journal encompasses genetic and functional genomic studies on human traits and on other model organisms, including mouse, fly, nematode and yeast. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation. According to the ''Journal Citation Reports'', the journal has a 2023 impact factor The impact factor (IF) or journal impact factor (JIF) of an academic journal is a type of journal ranking. Journals with higher impact factor values are considered more prestigious or important within their field. The Impact Factor of a journa ... of 31.7, ranking it 2nd out of 175 journals in the category "Genetics & Heredity". Further evaluation metrics from Scopus and Journal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, a Guanine, G nucleotide present at a specific location in a reference genome may be replaced by an Adenine, A in a minority of individuals. The two possible nucleotide variations of this SNP – G or A – are called alleles. SNPs can help explain differences in susceptibility to a wide range of diseases across a population. For example, a common SNP in the Factor H, CFH gene is associated with increased risk of age-related macular degeneration. Differences in the severity of an illness or response to treatments may also be manifestations of genetic variations caused by SNPs. For example, two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SNP Array
In molecular biology, SNP array is a type of DNA microarray which is used to detect polymorphisms within a population. A single nucleotide polymorphism (SNP), a variation at a single site in DNA, is the most frequent type of variation in the genome. Around 335 million SNPs have been identified in the human genome, 15 million of which are present at frequencies of 1% or higher across different populations worldwide. Principles The basic principles of SNP array are the same as the DNA microarray. These are the convergence of DNA hybridization, fluorescence microscopy, and solid surface DNA capture. The three mandatory components of the SNP arrays are: # An array containing immobilized allele-specific oligonucleotide (ASO) probes. # Fragmented nucleic acid sequences of target, labelled with fluorescent dyes. # A detection system that records and interprets the hybridization signal. The ASO probes are often chosen based on sequencing of a representative panel of individuals: pos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Identity By Descent
A DNA segment is identical by descent (IBD) in two or more individuals if: * they have inherited it from a common ancestor without recombination, that is, the segment has the same ancestral origin in these individuals * the segment is maximal, that is, it is delimited at both ends by ancestral recombination events. Theory All individuals in a finite population are related if traced back long enough and will, therefore, share segments of their genomes IBD. During meiosis segments of IBD are broken up by recombination. Therefore, the expected length of an IBD segment depends on the number of generations since the most recent common ancestor at the locus of the segment. The length of IBD segments that result from a common ancestor ''n'' generations in the past (therefore involving 2''n'' meiosis) is exponentially distributed with mean 1/(2''n'') Morgans (M). The expected number of IBD segments decreases as the number of generations since the common ancestor at this locus incr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microsatellite
A microsatellite is a tract of repetitive DNA in which certain Sequence motif, DNA motifs (ranging in length from one to six or more base pairs) are repeated, typically 5–50 times. Microsatellites occur at thousands of locations within an organism's genome. They have a higher mutation rate than other areas of DNA leading to high genetic diversity. Microsatellites are often referred to as short tandem repeats (STRs) by forensic genetics, forensic geneticists and in genetic genealogy, or as simple sequence repeats (SSRs) by plant geneticists. Microsatellites and their longer cousins, the minisatellites, together are classified as variable number tandem repeat, VNTR (variable number of tandem repeats) DNA. The name Satellite DNA, "satellite" DNA refers to the early observation that centrifugation of genomic DNA in a test tube separates a prominent layer of bulk DNA from accompanying "satellite" layers of repetitive DNA. They are widely used for DNA profiling in Loss of heterozygos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Íslendingabók (genealogical Database)
(, literally 'book of Icelanders') is a database created by the biotechnology company deCODE genetics and Friðrik Skúlason, attempting to record the genealogy of all Icelanders who have ever lived, where sources are available. takes its name from the Íslendingabók, first history of Iceland, commonly attributed to Ari the Wise. History Genealogy has been a pastime of Icelanders for centuries, with its roots in medieval political agenda. Texts from early ages of Icelandic history, containing genealogical information, have survived into the modern age and scholars and enthusiasts have maintained the genealogy knowledge through the ages. In early 1988, Friðrik Skúlason marketed a software program for registering family information and started to compile a database of Icelandic genealogy with the aim to register all available Icelandic genealogy information. In 1997, deCODE genetics and Mr. Skúlason entered into an agreement to speed up the compilation of the database and to ena ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Icelandic Census Of 1703
The Icelandic census of 1703 was the first census () of Iceland and the oldest complete census of any country that has survived. It was listed in UNESCO's Memory of the World international register in 2013. Procedure of the census The census recorded the name, age, residence, and social standing of every inhabitant; it was the first such complete census.Gunnar Karlsson, ''The History of Iceland'', Minneapolis: University of Minnesota, 2000, p. 162 Those without fixed address were recorded under the place where they spent the night before Easter.Richard F. Tomasson, ''Iceland, the First New Society'', Minneapolis: University of Minnesota, 1980, p. 72 The census was assembled and organized by two Icelanders, Árni Magnússon, who had just been appointed a professor at the University of Copenhagen, and Páll Vídalín, sheriff and vice-lawman. They were commissioned in 1702 by King Frederick IV of Denmark to perform a complete survey of Iceland, then a Danish possession, in order to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FRISK Software International
FRISK Software International (established in 1993) was an Icelandic software company that developed F-Prot antivirus and F-Prot AVES antivirus and anti-spam service. The company was founded in 1993. It was acquired by Cyren in 2012. History The company was founded in 1993. Its name is derived from the initial letters of the personal name and patronymic of ''Fri''ðrik ''Sk''úlason, its founder. Dr. Vesselin Vladimirov Bontchev, a computer expert from Bulgaria, best known for his research on the Dark Avenger virus, worked for the company as an anti-virus researcher. F-Prot Antivirus was first released in 1989, making it one of the longest lived anti-virus brands on the market. It was the world's first with a heuristic engine. It is sold in both home and corporate packages, of which there are editions for Windows and Linux. There are corporate versions for Microsoft Exchange, Solaris, and certain IBM eServers. The Linux version is available to home users free of charge, with ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |