|
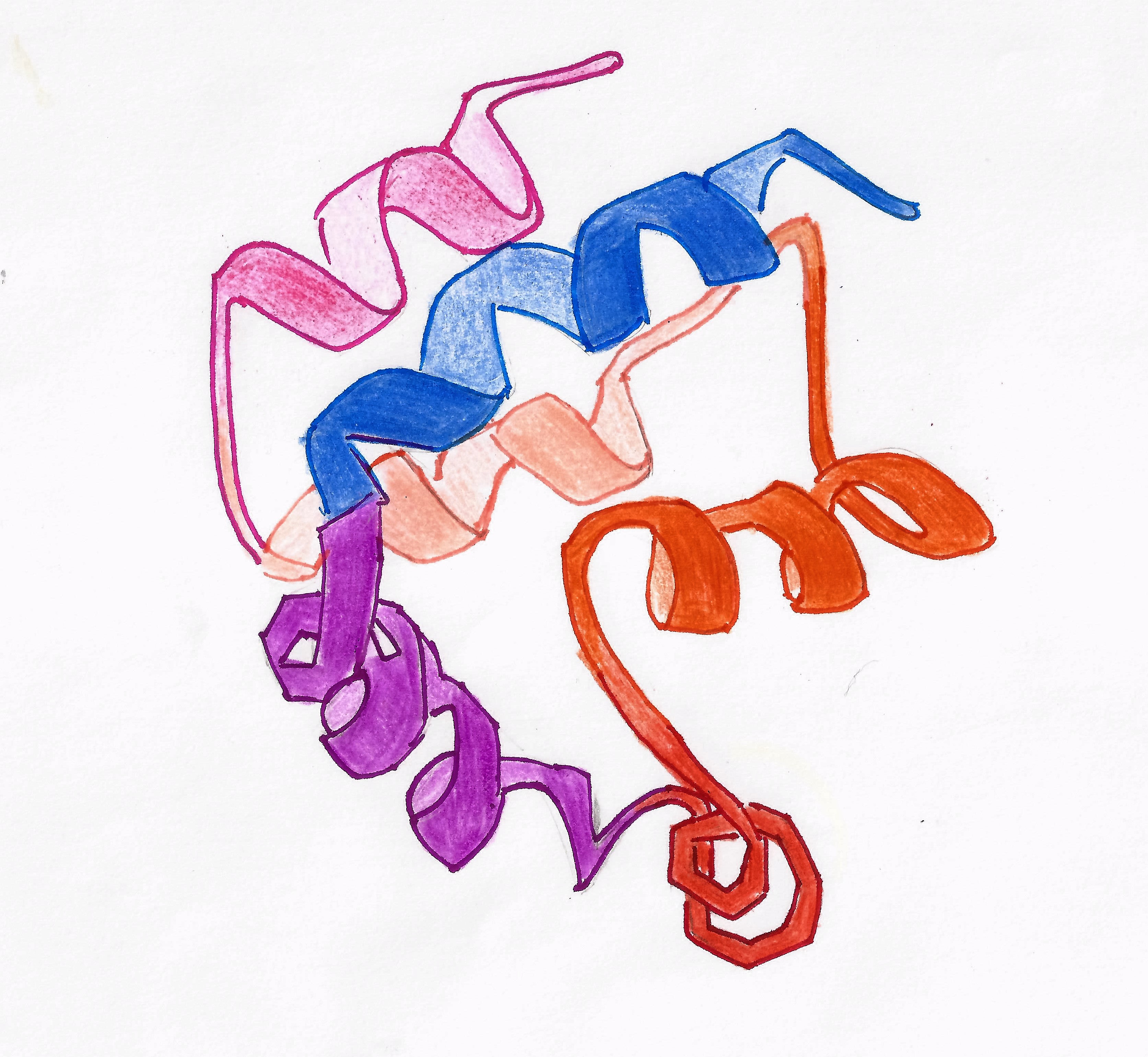
CD95
The Fas receptor, also known as Fas, FasR, apoptosis antigen 1 (APO-1 or APT), cluster of differentiation 95 (CD95) or tumor necrosis factor receptor superfamily member 6 (TNFRSF6), is a protein that in humans is encoded by the ''FAS'' gene. Fas was first identified using a monoclonal antibody generated by immunizing mice with the FS-7 cell line. Thus, the name Fas is derived from ''F''S-7-''a''ssociated ''s''urface antigen. The Fas receptor is a death receptor on the surface of cells that leads to programmed cell death (apoptosis) if it binds its ligand, Fas ligand (FasL). It is one of two apoptosis pathways, the other being the mitochondrial pathway. Gene FAS receptor gene is located on the long arm of chromosome 10 (10q24.1) in humans and on chromosome 19 in mice. The gene lies on the plus ( Watson strand) and is 25,255 bases in length organized into nine protein encoding exons. Similar sequences related by evolution (orthologs) are found in most mammals. Protein Prev ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Death-inducing Signaling Complex
The death-inducing signaling complex or DISC is a multi-protein complex formed by members of the death receptor family of apoptosis-inducing cellular receptors. A typical example is FasR, which forms the DISC upon trimerization as a result of its ligand ( FasL) binding. The DISC is composed of the death receptor, FADD, and caspase 8. It transduces a downstream signal cascade resulting in apoptosis. Description The Fas ligands, or cytotoxicity-dependent APO-1-associated proteins, physically associate with APO-1 (also known as the Fas receptor, or CD95), a tumor necrosis factor containing a functional death domain. This association leads to the formation of the DISC, thereby inducing apoptosis. The entire process is initiated when the cell registers the presence of CD95L, the cognate ligand for APO-1. Upon binding, the CAP proteins and procaspase-8 (composed of FLICE, MACH, and Mch5) bind to CD95 through death domain and death effector domain interactions. Procaspase-8 activa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Death Effector Domain
The death-effector domain (DED) is a protein interaction domain found only in eukaryotes that regulates a variety of cellular signalling pathways. The DED domain is found in inactive procaspases (cysteine proteases) and proteins that regulate caspase activation in the apoptosis cascade such as FAS-associating death domain-containing protein (FADD). FADD recruits procaspase 8 and procaspase 10 into a death induced signaling complex (DISC). This recruitment is mediated by a homotypic interaction between the procaspase DED and a second DED that is death effector domain in an adaptor protein that is directly associated with activated TNF receptors. Complex formation allows proteolytic activation of procaspase into the active caspase form which results in the initiation of apoptosis (cell death). Structurally the DED domain are a subclass of protein motif known as the death fold and contains 6 alpha helices, that closely resemble the structure of the Death domain (DD). Structure ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FADD
FAS-associated death domain protein, also called MORT1, is encoded by the ''FADD'' gene on the 11q13.3 region of chromosome 11 in humans. FADD is an adaptor protein that bridges members of the tumor necrosis factor receptor superfamily, such as the Fas-receptor, to procaspases 8 and 10 to form the death-inducing signaling complex (DISC) during apoptosis. As well as its most well known role in apoptosis, FADD has also been seen to play a role in other processes including proliferation, cell cycle regulation and development. Structure FADD is a 23 kDa protein, made up of 208 amino acids. It contains two main domains: a C terminal death domain (DD) and an N terminal death effector domain (DED). Each domain, although sharing very little sequence similarity, are structurally similar to one another, with each consisting of 6 α helices. The DD of FADD binds to receptors such as the Fas receptor at the plasma membrane via their DD. The interaction between the death domains are e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apoptosis
Apoptosis (from grc, ἀπόπτωσις, apóptōsis, 'falling off') is a form of programmed cell death that occurs in multicellular organisms. Biochemical events lead to characteristic cell changes ( morphology) and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, DNA fragmentation, and mRNA decay. The average adult human loses between 50 and 70 billion cells each day due to apoptosis. For an average human child between eight and fourteen years old, approximately twenty to thirty billion cells die per day. In contrast to necrosis, which is a form of traumatic cell death that results from acute cellular injury, apoptosis is a highly regulated and controlled process that confers advantages during an organism's life cycle. For example, the separation of fingers and toes in a developing human embryo occurs because cells between the digits undergo apoptosis. Unlike necrosis, apoptosis produces cell fragments called apop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alternative Splicing
Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tumor Suppressor
A tumor suppressor gene (TSG), or anti-oncogene, is a gene that regulates a cell during cell division and replication. If the cell grows uncontrollably, it will result in cancer. When a tumor suppressor gene is mutated, it results in a loss or reduction in its function. In combination with other genetic mutations, this could allow the cell to grow abnormally. The loss of function for these genes may be even more significant in the development of human cancers, compared to the activation of oncogenes. TSGs can be grouped into the following categories: caretaker genes, gatekeeper genes, and more recently landscaper genes. Caretaker genes ensure stability of the genome via DNA repair and subsequently when mutated allow mutations to accumulate. Meanwhile, gatekeeper genes directly regulate cell growth by either inhibiting cell cycle progression or inducing apoptosis. Lastly landscaper genes regulate growth by contributing to the surrounding environment, when mutated can cause an env ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tumor Necrosis Factor Receptor Superfamily
The tumor necrosis factor receptor superfamily (TNFRSF) is a protein superfamily of cytokine receptors characterized by the ability to bind tumor necrosis factors (TNFs) via an extracellular cysteine-rich domain. With the exception of nerve growth factor (NGF), all TNFs are homologous to the archetypal TNF-alpha. In their active form, the majority of TNF receptors form trimeric complexes in the plasma membrane. Accordingly, most TNF receptors contain transmembrane domains (TMDs), although some can be cleaved into soluble forms (e.g. TNFR1), and some lack a TMD entirely (e.g. DcR3). In addition, most TNF receptors require specific adaptor protein such as TRADD, TRAF, RIP and FADD for downstream signalling. TNF receptors are primarily involved in apoptosis and inflammation, but they can also take part in other signal transduction pathways, such as proliferation, survival, and differentiation. TNF receptors are expressed in a wide variety of tissues in mammals, especially in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tumor Necrosis Factor Receptor
The tumor necrosis factor receptor superfamily (TNFRSF) is a protein superfamily of cytokine receptors characterized by the ability to bind tumor necrosis factors (TNFs) via an extracellular cysteine-rich domain. With the exception of nerve growth factor (NGF), all TNFs are homologous to the archetypal TNF-alpha. In their active form, the majority of TNF receptors form trimeric complexes in the plasma membrane. Accordingly, most TNF receptors contain transmembrane domains (TMDs), although some can be cleaved into soluble forms (e.g. TNFR1), and some lack a TMD entirely (e.g. DcR3). In addition, most TNF receptors require specific adaptor protein such as TRADD, TRAF, RIP and FADD for downstream signalling. TNF receptors are primarily involved in apoptosis and inflammation, but they can also take part in other signal transduction pathways, such as proliferation, survival, and differentiation. TNF receptors are expressed in a wide variety of tissues in mammals, especially i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Icahn School Of Medicine At Mount Sinai
The Icahn School of Medicine at Mount Sinai (ISMMS or Mount Sinai), formerly the Mount Sinai School of Medicine, is a private medical school in New York City. It is the academic teaching arm of the Mount Sinai Health System, which manages eight hospital campuses in the New York metropolitan area, including Mount Sinai Hospital and the New York Eye and Ear Infirmary. Mount Sinai is ranked #11 among American medical schools by the 2023 '' U.S. News & World Report''.https://www.usnews.com/best-graduate-schools/top-medical-schools/icahn-school-of-medicine-at-mount-sinai-04072 In 2021, it was ranked 15th in the country for biomedical research and leads the country in research funding from the National Institutes of Health for neuroscience (#2) and genetics (#2). It attracted over $400 million in total NIH funding in 2021. Mount Sinai's faculty includes 23 elected members of the National Academies of Sciences, Engineering, and Medicine and 40 members of the American Society fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
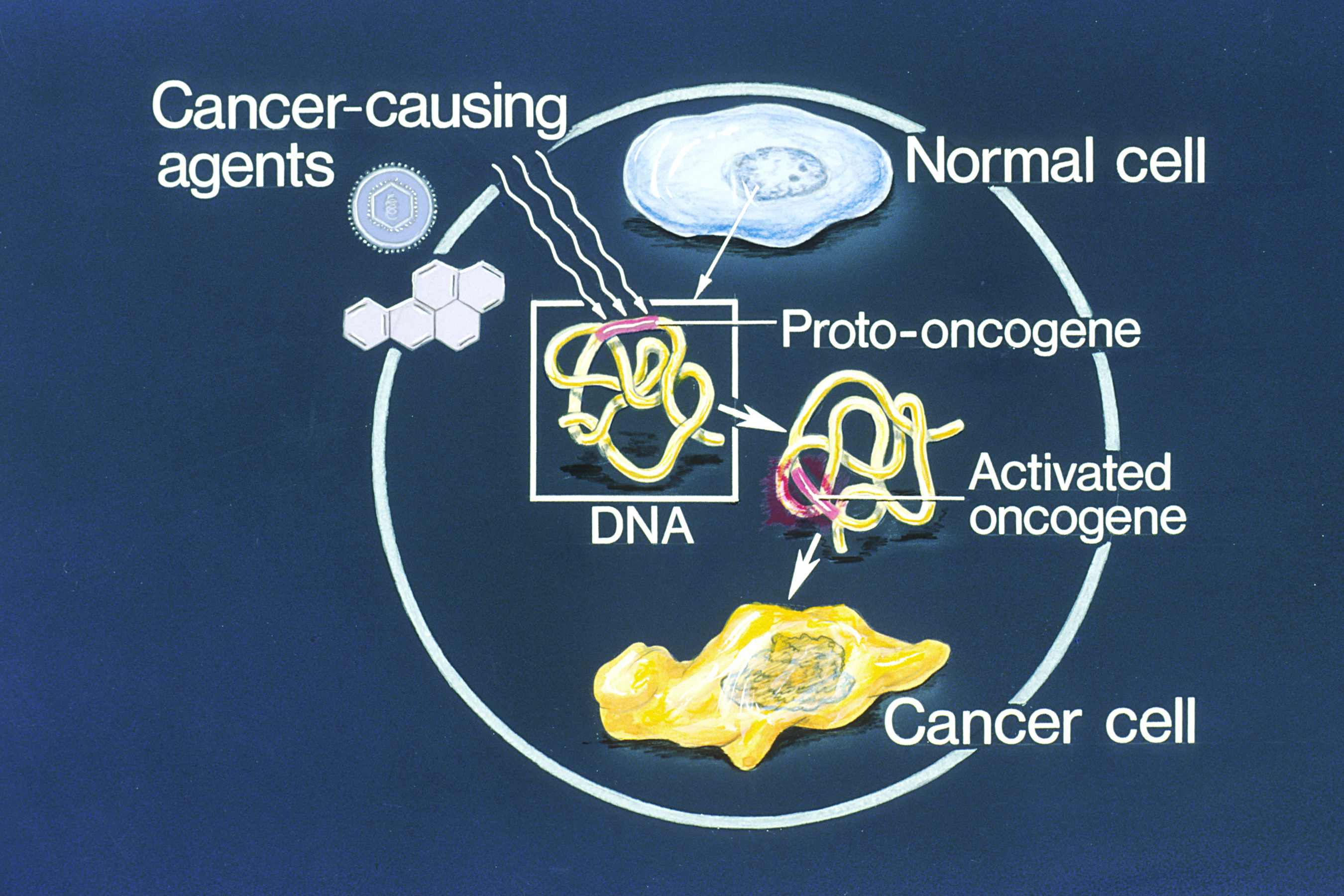
Oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.Kimball's Biology Pages. "Oncogenes" Free full text Most normal cells will undergo a programmed form of rapid cell death () when critical functions are altered and malfunctioning. Activated oncogenes can cause those cells designated for apoptosis to survive and proliferate instead. Most oncogenes began as proto-oncogenes: normal genes involved in cell growth and proliferation or inhibition of apoptosis. If, through mutation, normal genes promoting cellular growth are up-regulated (gain-of-function mutation), they will predis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteolytic Cleavage
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion. Proteolysis in organisms serves many purposes; for example, digestive enzymes break down proteins in food to provide amino acids for the organism, while proteolytic processing of a polypeptide chain after its synthesis may be necessary for the production of an active protein. It is also important in the regulation of some physiological and cellular processes including apoptosis, as well as preventing the accumulation of unwanted or misfolded proteins in cells. Consequently, abnormality in the regulation of proteolysis can cause disease. Proteolysis can also be used as an analytical tool for studying proteins in the laboratory, and it may also be used in industry, for example in food proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |