|
Catalytic RNA
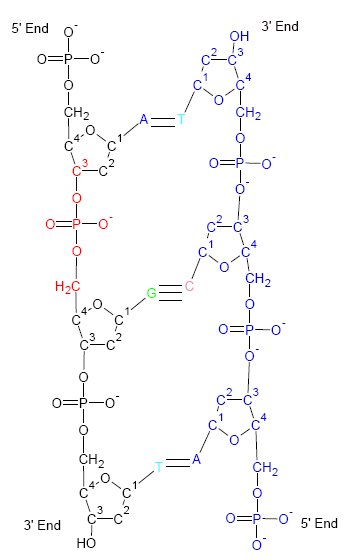
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or ''in vitro'' evolved ribozymes are the cleavage (or ligation) of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3') can aminoacylate a GCCU-3' sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during protein synthesis. They also participate in a variety of RNA processing reactions, including RNA splicing, viral replicati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Full Length Hammerhead Ribozyme
{{disambiguatio ...
Full may refer to: * People with the surname Full, including: ** Mr. Full (given name unknown), acting Governor of German Cameroon, 1913 to 1914 * A property in the mathematical field of topology; see Full set * A property of functors in the mathematical field of category theory; see Full and faithful functors * Satiety, the absence of hunger * A standard bed size, see Bed * Full house (poker), a type of poker hand * Fulling, also known as tucking or walking ("waulking" in Scotland), term for a step in woollen clothmaking (verb: ''to full'') * Full-Reuenthal, a municipality in the district of Zurzach in the canton of Aargau in Switzerland See also *"Fullest", a song by the rapper Cupcakke *Ful (other) Ful or FUL may refer to: * Fula language * Fula people * Ful medames, a fava bean dish of Sudan and Egypt * Fullerton Municipal Airport, California, United States; IATA code FUL * Fullerton Transportation Center The Fullerton Transportation Ce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hairpin Ribozyme
The hairpin ribozyme is a small section of RNA that can act as a ribozyme. Like the hammerhead ribozyme it is found in RNA satellite (biology), satellites of plant viruses. It was first identified in the minus strand of the tobacco ringspot virus (TRSV) satellite RNA where it Catalysis, catalyzes self-cleavage and joining (Chemical ligation, ligation) Chemical reaction, reactions to process the products of Rolling circle replication, rolling circle virus replication into linear and circular satellite RNA molecules. The hairpin ribozyme is similar to the hammerhead ribozyme in that it does not require a metal ion for the reaction. Biological function The hairpin ribozyme is an RNA motif that catalyzes RNA processing reactions essential for replication of the satellite RNA molecules in which it is embedded. These reactions are self-processing, i.e. a molecule rearranging its own structure. Both cleavage and end joining reactions are mediated by the ribozyme motif, leading to a mi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gene... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978) The term ''intron'' refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons. Introns are found in the genes of most eukaryotes and many eukaryotic viruses, and they can be located in both protein-coding genes and genes that function as RNA ( noncoding genes). There are four main types of introns: tRNA introns, group I introns, group I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Leslie Orgel
Leslie Eleazer Orgel FRS (12 January 1927 – 27 October 2007) was a British chemist and member of the National Academy of Sciences, known for his theories on the origin of life. Biography Leslie Orgel was born in London on . He received his Bachelor of Arts degree in chemistry with first-class honours from the University of Oxford in 1948. In 1951 he was elected a Fellow of Magdalen College, Oxford and in 1953 was awarded his PhD in chemistry. Orgel started his career as a theoretical inorganic chemist and continued his studies in this field at Oxford, the California Institute of Technology, and the University of Chicago. Together with Sydney Brenner, Jack Dunitz, Dorothy Hodgkin, and Beryl M. Oughton he was one of the first people in April 1953 to see the model of the structure of DNA, constructed by Francis Crick and James Watson, at the time he and the other scientists were working at Oxford University's Chemistry Department. According to the late Dr. Beryl Ought ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Francis Crick
Francis Harry Compton Crick (8 June 1916 – 28 July 2004) was an English molecular biologist, biophysicist, and neuroscientist. He, James Watson, Rosalind Franklin, and Maurice Wilkins played crucial roles in deciphering the Nucleic acid double helix, helical structure of the DNA molecule. Crick and Watson's paper in ''Nature (journal), Nature'' in 1953 laid the groundwork for understanding DNA structure and functions. Together with Maurice Wilkins, they were jointly awarded the 1962 Nobel Prize in Physiology or Medicine "for their discoveries concerning the molecular geometry, molecular structure of nucleic acids and its significance for information transfer in living material". Crick was an important theoretical molecular biologist and played a crucial role in research related to revealing the helical structure of DNA. He is widely known for the use of the term "central dogma" to summarise the idea that once information is transferred from nucleic acids (DNA or RNA) to prot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carl Woese
Carl Richard Woese ( ; July 15, 1928 – December 30, 2012) was an American microbiologist and biophysicist. Woese is famous for defining the Archaea (a new domain of life) in 1977 through a pioneering phylogenetic taxonomy of 16S ribosomal RNA, a technique that has revolutionized microbiology. He also originated the RNA world hypothesis in 1967, although not by that name. Woese held the Stanley O. Ikenberry Chair and was professor of microbiology at the University of Illinois Urbana–Champaign. Life and education Woese was born in Syracuse, New York on July 15, 1928. His family was German American. Woese attended Deerfield Academy in Massachusetts. He received a bachelor's degree in mathematics and physics from Amherst College in 1950. During his time at Amherst, Woese took only one biology course (Biochemistry, in his senior year) and had "no scientific interest in plants and animals" until advised by William M. Fairbank, then an assistant professor of physics at A ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalysis
Catalysis () is the increase in rate of a chemical reaction due to an added substance known as a catalyst (). Catalysts are not consumed by the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quickly, very small amounts of catalyst often suffice; mixing, surface area, and temperature are important factors in reaction rate. Catalysts generally react with one or more reactants to form intermediates that subsequently give the final reaction product, in the process of regenerating the catalyst. The rate increase occurs because the catalyst allows the reaction to occur by an alternative mechanism which may be much faster than the noncatalyzed mechanism. However the noncatalyzed mechanism does remain possible, so that the total rate (catalyzed plus noncatalyzed) can only increase in the presence of the catalyst and never decrease. Catalysis may be classified as either homogeneous, whose components are dispersed in the same phase (usual ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metabolic reactions, DNA replication, Cell signaling, responding to stimuli, providing Cytoskeleton, structure to cells and Fibrous protein, organisms, and Intracellular transport, transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the Nucleic acid sequence, nucleotide sequence of their genes, and which usually results in protein folding into a specific Protein structure, 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called pep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to Catalysis, catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or Directed evolution, ''in vitro'' evolved ribozymes are the cleavage (or Ligation (molecular biology), ligation) of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3') can aminoacylate a GCCU-3' sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during Translation (biology), protein synthesis. They also participate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Functional Genomics
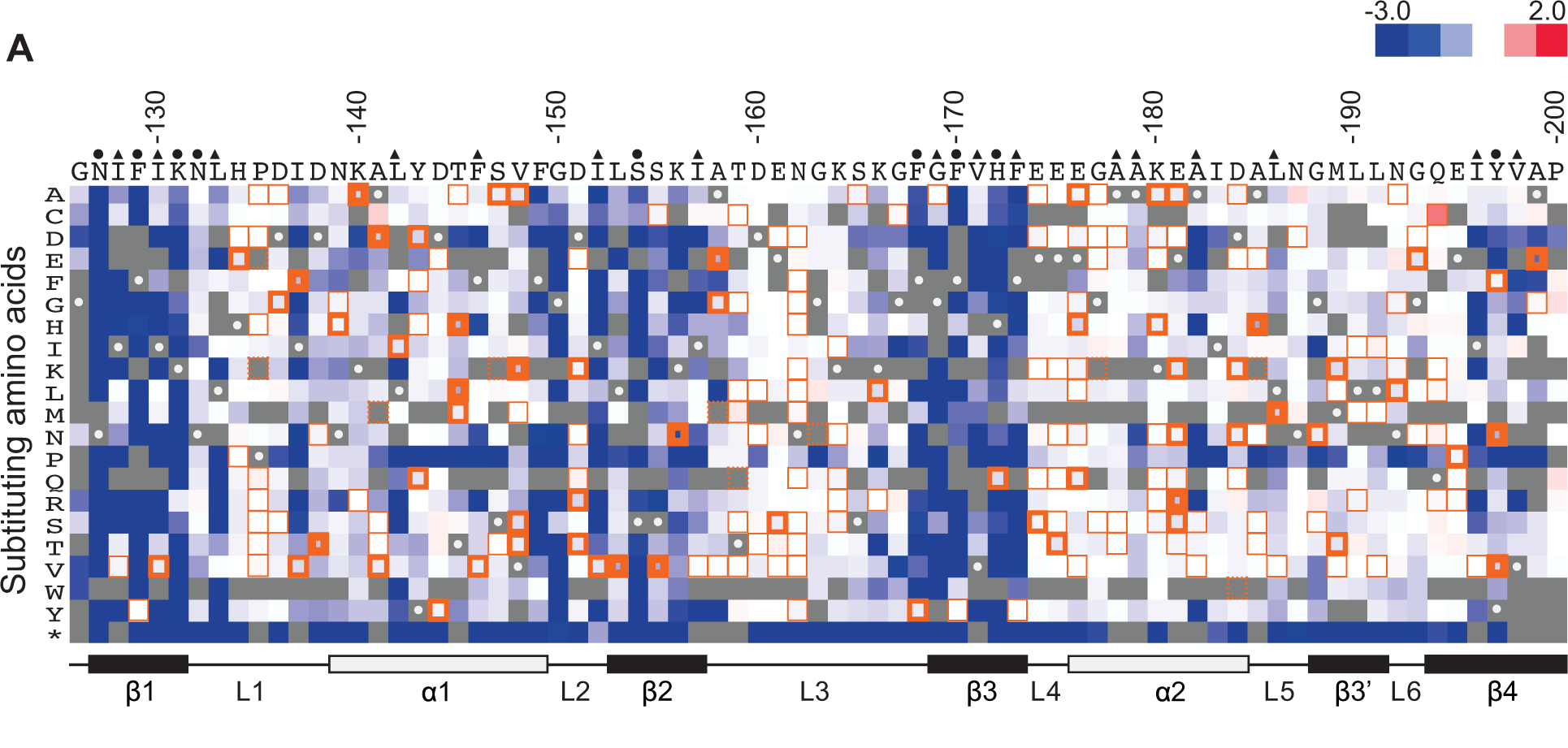
Functional genomics is a field of molecular biology that attempts to describe gene (and protein) functions and interactions. Functional genomics make use of the vast data generated by genomic and transcriptomic projects (such as genome sequencing projects and RNA sequencing). Functional genomics focuses on the dynamic aspects such as gene transcription, translation, regulation of gene expression and protein–protein interactions, as opposed to the static aspects of the genomic information such as DNA sequence or structures. A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional "candidate-gene" approach. Definition and goals In order to understand functional genomics it is important to first define function. In their paper Graur et al. define function in two possible ways. These are "selected effect" and "causal role". The "selected effect" function re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |