|
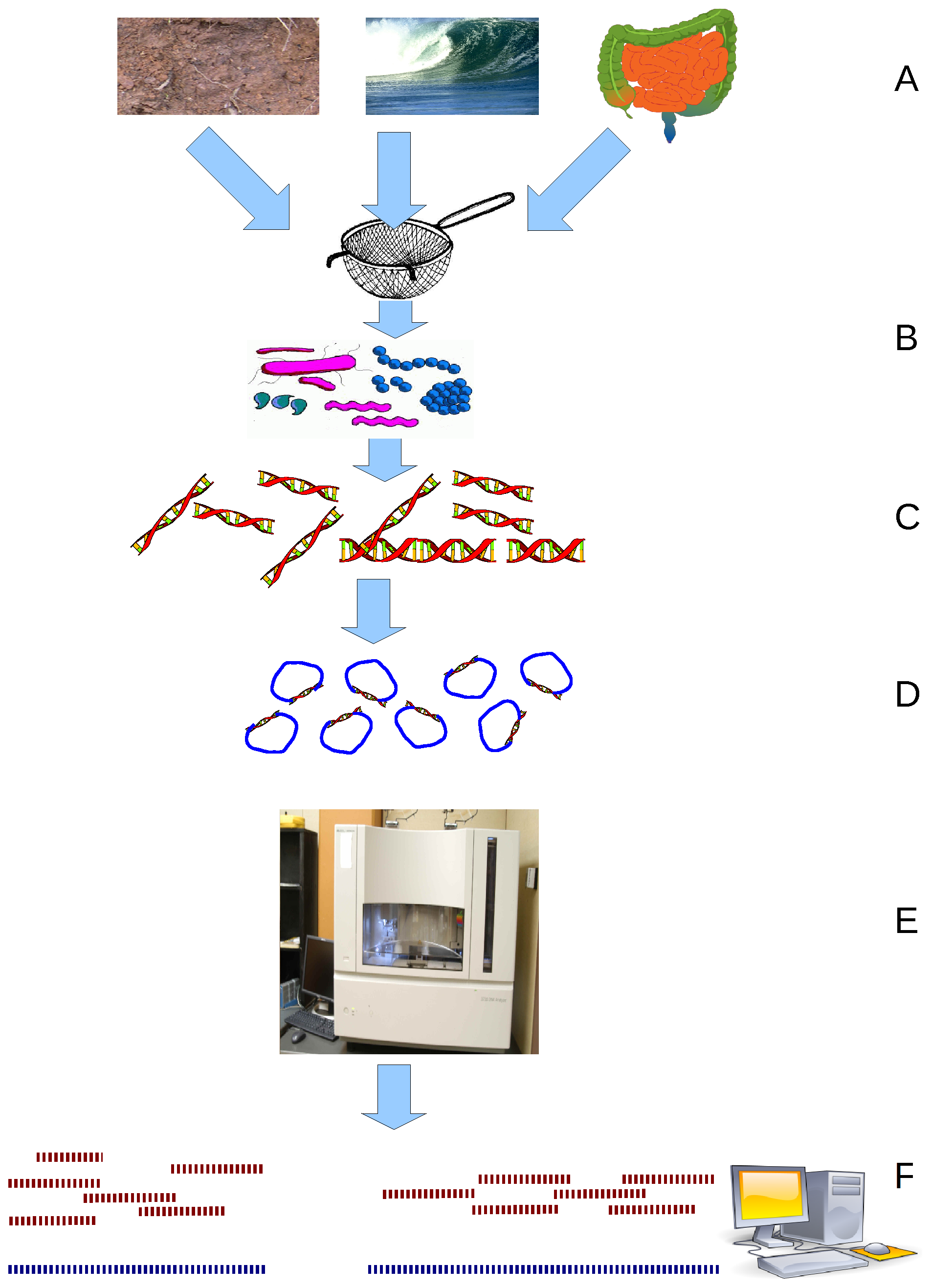
Binning (Metagenomics)
In metagenomics, binning is the process of grouping reads or contigs and assigning them to individual genome. Binning methods can be based on either compositional features or alignment (similarity), or both. Introduction Metagenomic samples can contain reads from a huge number of organisms. For example, in a single gram of soil, there can be up to 18000 different types of organisms, each with its own genome. Metagenomic studies sample DNA from the whole community, and make it available as nucleotide sequences of certain length. In most cases, the incomplete nature of the obtained sequences makes it hard to assemble individual genes, much less recovering the full genomes of each organism. Thus, binning techniques represent a "best effort" to identify reads or contigs within certain genome known as Metagenome Assembled Genome (MAG). Taxonomy of MAGs can be inferred through placement into reference phylogenetic tree using algorithms like GTDB-Tk. The first studies that sampled DN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metagenomics
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater sc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences, such as calculating the distance cost between strings in a natural language or in financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence alignments of proteins, the degree of similarity between amino acids occupying a p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
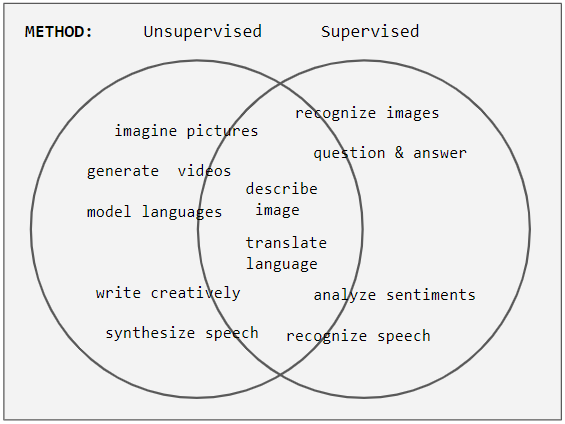
Supervised Learning
Supervised learning (SL) is a machine learning paradigm for problems where the available data consists of labelled examples, meaning that each data point contains features (covariates) and an associated label. The goal of supervised learning algorithms is learning a function that maps feature vectors (inputs) to labels (output), based on example input-output pairs. It infers a function from ' consisting of a set of ''training examples''. In supervised learning, each example is a ''pair'' consisting of an input object (typically a vector) and a desired output value (also called the ''supervisory signal''). A supervised learning algorithm analyzes the training data and produces an inferred function, which can be used for mapping new examples. An optimal scenario will allow for the algorithm to correctly determine the class labels for unseen instances. This requires the learning algorithm to generalize from the training data to unseen situations in a "reasonable" way (see inductive ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hierarchical Classification
Hierarchical classification is a system of grouping things according to a hierarchy. In the field of machine learning, hierarchical classification is sometimes referred to as instance space decomposition, which splits a complete multi-class problem into a set of smaller classification problems. See also * Deductive classifier * Cascading classifiers * Faceted classification A faceted classification is a classification scheme used in organizing knowledge into a systematic order. A faceted classification uses semantic categories, either general or subject-specific, that are combined to create the full classification ent ... References External links Hierarchical Classification – a useful approach for predicting thousands of possible categories Classification algorithms {{AI-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Support Vector Machine
In machine learning, support vector machines (SVMs, also support vector networks) are supervised learning models with associated learning algorithms that analyze data for classification and regression analysis. Developed at AT&T Bell Laboratories by Vladimir Vapnik with colleagues (Boser et al., 1992, Guyon et al., 1993, Cortes and Vapnik, 1995, Vapnik et al., 1997) SVMs are one of the most robust prediction methods, being based on statistical learning frameworks or VC theory proposed by Vapnik (1982, 1995) and Chervonenkis (1974). Given a set of training examples, each marked as belonging to one of two categories, an SVM training algorithm builds a model that assigns new examples to one category or the other, making it a non- probabilistic binary linear classifier (although methods such as Platt scaling exist to use SVM in a probabilistic classification setting). SVM maps training examples to points in space so as to maximise the width of the gap between the two categorie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Standard Score
In statistics, the standard score is the number of standard deviations by which the value of a raw score (i.e., an observed value or data point) is above or below the mean value of what is being observed or measured. Raw scores above the mean have positive standard scores, while those below the mean have negative standard scores. It is calculated by subtracting the population mean from an individual raw score and then dividing the difference by the population standard deviation. This process of converting a raw score into a standard score is called standardizing or normalizing (however, "normalizing" can refer to many types of ratios; see normalization for more). Standard scores are most commonly called ''z''-scores; the two terms may be used interchangeably, as they are in this article. Other equivalent terms in use include z-values, normal scores, standardized variables and pull in high energy physics. Computing a z-score requires knowledge of the mean and standard d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GC-content
In molecular biology and genetics, GC-content (or guanine-cytosine content) is the percentage of nitrogenous bases in a DNA or RNA molecule that are either guanine (G) or cytosine (C). This measure indicates the proportion of G and C bases out of an implied four total bases, also including adenine and thymine in DNA and adenine and uracil in RNA. GC-content may be given for a certain fragment of DNA or RNA or for an entire genome. When it refers to a fragment, it may denote the GC-content of an individual gene or section of a gene (domain), a group of genes or gene clusters, a non-coding region, or a synthetic oligonucleotide such as a primer. Structure Qualitatively, guanine (G) and cytosine (C) undergo a specific hydrogen bonding with each other, whereas adenine (A) bonds specifically with thymine (T) in DNA and with uracil (U) in RNA. Quantitatively, each GC base pair is held together by three hydrogen bonds, while AT and AU base pairs are held together by two hydrogen bon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biological Databases
Biological databases are libraries of biological sciences, collected from scientific experiments, published literature, high-throughput experiment technology, and computational analysis. They contain information from research areas including genomics, proteomics, metabolomics, microarray gene expression, and phylogenetics. Information contained in biological databases includes gene function, structure, localization (both cellular and chromosomal), clinical effects of mutations as well as similarities of biological sequences and structures. Biological databases can be classified by the kind of data they collect (see below). Broadly, there are molecular databases (for sequences, molecules, etc.), functional databases (for physiology, enzyme activities, phenotypes, ecology etc), taxonomic databases (for species and other taxonomic ranks), images and other media, or specimens (for museum collections etc.) Databases are important tools in assisting scientists to analyze and explain a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Unsupervised Learning
Unsupervised learning is a type of algorithm that learns patterns from untagged data. The hope is that through mimicry, which is an important mode of learning in people, the machine is forced to build a concise representation of its world and then generate imaginative content from it. In contrast to supervised learning where data is tagged by an expert, e.g. tagged as a "ball" or "fish", unsupervised methods exhibit self-organization that captures patterns as probability densities or a combination of neural feature preferences encoded in the machine's weights and activations. The other levels in the supervision spectrum are reinforcement learning where the machine is given only a numerical performance score as guidance, and semi-supervised learning where a small portion of the data is tagged. Neural networks Tasks vs. methods Neural network tasks are often categorized as discriminative (recognition) or generative (imagination). Often but not always, discriminative tas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
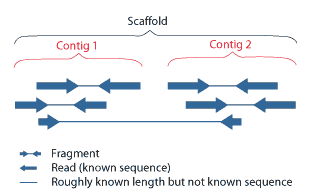
Contig
A contig (from ''contiguous'') is a set of overlapping DNA segments that together represent a consensus region of DNA.Gregory, S. ''Contig Assembly''. Encyclopedia of Life Sciences, 2005. In bottom-up sequencing projects, a contig refers to overlapping sequence data ( reads); in top-down sequencing projects, contig refers to the overlapping clones that form a physical map of the genome that is used to guide sequencing and assembly.Dear, P. H. ''Genome Mapping''. Encyclopedia of Life Sciences, 2005. . Contigs can thus refer both to overlapping DNA sequences and to overlapping physical segments (fragments) contained in clones depending on the context. Original definition of contig In 1980, Staden wrote: ''In order to make it easier to talk about our data gained by the shotgun method of sequencing we have invented the word "contig". A contig is a set of gel readings that are related to one another by overlap of their sequences. All gel readings belong to one and only one con ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Supervised Learning
Supervised learning (SL) is a machine learning paradigm for problems where the available data consists of labelled examples, meaning that each data point contains features (covariates) and an associated label. The goal of supervised learning algorithms is learning a function that maps feature vectors (inputs) to labels (output), based on example input-output pairs. It infers a function from ' consisting of a set of ''training examples''. In supervised learning, each example is a ''pair'' consisting of an input object (typically a vector) and a desired output value (also called the ''supervisory signal''). A supervised learning algorithm analyzes the training data and produces an inferred function, which can be used for mapping new examples. An optimal scenario will allow for the algorithm to correctly determine the class labels for unseen instances. This requires the learning algorithm to generalize from the training data to unseen situations in a "reasonable" way (see inductive ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebacteria kingdom), but this term has fallen out of use. Archaeal cells have unique properties separating them from the other two domains, Bacteria and Eukaryota. Archaea are further divided into multiple recognized phyla. Classification is difficult because most have not been isolated in a laboratory and have been detected only by their gene sequences in environmental samples. Archaea and bacteria are generally similar in size and shape, although a few archaea have very different shapes, such as the flat, square cells of '' Haloquadratum walsbyi''. Despite this morphological similarity to bacteria, archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes, notably for the enzymes invo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


.jpg)