|
AsponA Antisense RNA
AsponA is a small asRNA transcribed antisense to the penicillin-binding protein 1A gene called ''ponA.'' It was identified by RNAseq and the expression was validated by 5' and 3' RACE experiments in '' Pseudomanas aeruginosa''. ''AsponA'' expression was up or down regulated under different antibiotic stress. Owing to its location it may be able to prevent the transcription or translation of the opposite gene. Study by Wurtzel ''et al.'' and Ferrara ''et al.'' also detected its expression. See also * NrsZ small RNA NrsZ (nitrogen regulated small RNA) is a bacterial small RNA found in the opportunistic pathogen ''Pseudomonas aeruginosa'' PAO1. Its transcription is induced during nitrogen limitation by the NtrB/C two-component system (an important regulator of ... * SrbA sRNA * ''Pseudomonas'' sRNA References {{Reflist Non-coding RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952. Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. Types The most common seconda ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Conservation
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, promp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, Hot spring, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the nitrogen fixation, fixation of nitrogen from the Earth's atmosphere, atmosphere. The nutrient cycle includes the decomposition of cadaver, dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antisense RNA
Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its translation into protein. asRNAs (which occur naturally) have been found in both prokaryotes and eukaryotes, and can be classified into short (200 nucleotides) non-coding RNAs (ncRNAs). The primary function of asRNA is regulating gene expression. asRNAs may also be produced synthetically and have found wide spread use as research tools for gene knockdown. They may also have therapeutic applications. Discovery and history in drug development Some of the earliest asRNAs were discovered while investigating functional proteins. An example was micF asRNA. While characterizing the outer membrane porin in ''E.coli'', some of the promoter clones observed were capable of repressing the expression of other membrane porin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rapid Amplification Of CDNA Ends
Rapid amplification of cDNA ends (RACE) is a technique used in molecular biology to obtain the full length sequence of an RNA transcript found within a cell. RACE results in the production of a cDNA copy of the RNA sequence of interest, produced through reverse transcription, followed by PCR amplification of the cDNA copies (see RT-PCR). The amplified cDNA copies are then sequenced and, if long enough, should map to a unique genomic region. RACE is commonly followed up by cloning before sequencing of what was originally individual RNA molecules. A more high-throughput alternative which is useful for identification of novel transcript structures, is to sequence the RACE-products by next generation sequencing technologies. Process RACE can provide the sequence of an RNA transcript from a small known sequence within the transcript to the 5' end (5' RACE-PCR) or 3' end (3' RACE-PCR) of the RNA. This technique is sometimes called ''one-sided PCR'' or ''anchored PCR''. The first step ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
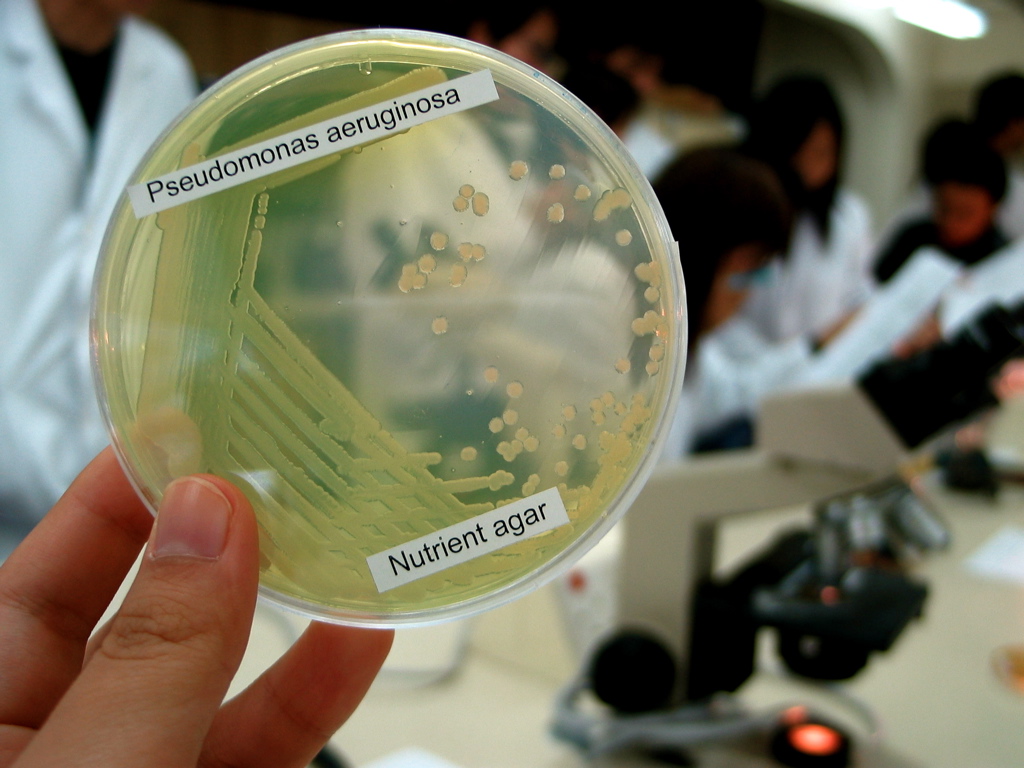
Pseudomonas Aeruginosa
''Pseudomonas aeruginosa'' is a common encapsulated, gram-negative, aerobic–facultatively anaerobic, rod-shaped bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, ''P. aeruginosa'' is a multidrug resistant pathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes. The organism is considered opportunistic insofar as serious infection often occurs during existing diseases or conditions – most notably cystic fibrosis and traumatic burns. It generally affects the immunocompromised but can also infect the immunocompetent as in hot tub folliculitis. Treatment of ''P. aeruginosa'' infections can be difficult due to its natural resistance to antibiotics. When more advanced antibiotic drug regimens are needed adverse effects ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NrsZ Small RNA
NrsZ (nitrogen regulated small RNA) is a bacterial small RNA found in the opportunistic pathogen ''Pseudomonas aeruginosa'' PAO1. Its transcription is induced during nitrogen limitation by the NtrB/C two-component system (an important regulator of nitrogen assimilation and swarming motility) together with the alternative sigma factor RpoN ( a global regulator involved in nitrogen metabolism). NrsZ by activating ''rhlA (''a gene essential for rhamnolipids synthesis) positively regulates the production of rhamnolipid Rhamnolipids are a class of glycolipid produced by ''Pseudomonas aeruginosa'', amongst other organisms, frequently cited as bacterial surfactants. They have a glycosyl head group, in this case a rhamnose moiety, and a 3-(hydroxyalkanoyloxy)alkanoic ... surfactants needed for swarming motility. See also * ''Pseudomonas'' sRNA * SrbA sRNA * AsponA antisense RNA References Non-coding RNA {{molecular-cell-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SrbA SRNA
SrbA ( sRNA regulator of biofilms A) is a small regulatory non-coding RNA identified in pathogenic ''Pseudomonas aeruginosa'.'' It is important for biofilm formation and pathogenicity. Bacterial strain with deleted SrbA had reduced biofilm mass. As the ability to form biofilms can contribute to the ability a pathogen to thrive within the host, the ''C. elegans'' hosts infected with the srbA deleted strain displayed significantly lower mortality rate than the wild-type strain. However, the deletion of srbA had no effect on growth or antibiotic resistance in ''P. aeruginosa''. See also * NrsZ small RNA * ''Pseudomonas'' sRNA * AsponA antisense RNA AsponA is a small asRNA transcribed antisense to the penicillin-binding protein 1A gene called ''ponA.'' It was identified by RNAseq and the expression was validated by 5' and 3' RACE experiments in '' Pseudomanas aeruginosa''. ''AsponA'' express ... References Non-coding RNA {{molecular-cell-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudomonas SRNA
''Pseudomonas'' sRNA are non-coding RNAs (ncRNA) that were predicted by the bioinformatics, bioinformatic program SRNApredict2. This program identifies putative sRNAs by searching for co-localization of genetic features commonly associated with sRNA-encoding genes and the gene expression, expression of the predicted sRNAs was subsequently confirmed by Northern blot analysis. These sRNAs have been shown to be conservation (genetics), conserved across several ''pseudomonas'' species but their function is yet to be determined. Using Tet-Trap genetic approach RNA thermometer, RNAT genes post-transcriptionally regulated by temperature upshift were identified: ''ptxS'' (implicated in virulence) and PA5194. See also *Bacillus subtilis BSR sRNAs, ''Bacillus subtilis'' sRNA *C. elegans small RNAs, ''Caenorhabditis elegans'' sRNA *Mycobacterium tuberculosis sRNA, ''Mycobacterium tuberculosis'' sRNA *Bacteroides thetaiotaomicron sRNA, ''Bacteroides thetaiotaomicron'' sRNA *NrsZ small RNA *Aspo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


_.png)