Variant Call Format on:
[Wikipedia]
[Google]
[Amazon]
 The Variant Call Format (VCF) specifies the format of a text file used in
The Variant Call Format (VCF) specifies the format of a text file used in
##phasing=partial
##INFO=
##INFO=
##INFO=
##INFO=
##INFO=
##INFO=
##FILTER=
##FILTER=
##FORMAT=
##FORMAT=
##FORMAT=
##FORMAT=
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA00001 NA00002 NA00003
20 14370 rs6054257 G A 29 PASS NS=3;DP=14;AF=0.5;DB;H2 GT:GQ:DP:HQ 0, 0:48:1:51,51 1, 0:48:8:51,51 1/1:43:5:.,.
20 17330 . T A 3 q10 NS=3;DP=11;AF=0.017 GT:GQ:DP:HQ 0, 0:49:3:58,50 0, 1:3:5:65,3 0/0:41:3
20 1110696 rs6040355 A G,T 67 PASS NS=2;DP=10;AF=0.333,0.667;AA=T;DB GT:GQ:DP:HQ 1, 2:21:6:23,27 2, 1:2:0:18,2 2/2:35:4
20 1230237 . T . 47 PASS NS=3;DP=13;AA=T GT:GQ:DP:HQ 0, 0:54:7:56,60 0, 0:48:4:51,51 0/0:61:2
20 1234567 microsat1 GTC G,GTCT 50 PASS NS=3;DP=9;AA=G GT:GQ:DP 0/1:35:4 0/2:17:2 1/1:40:3
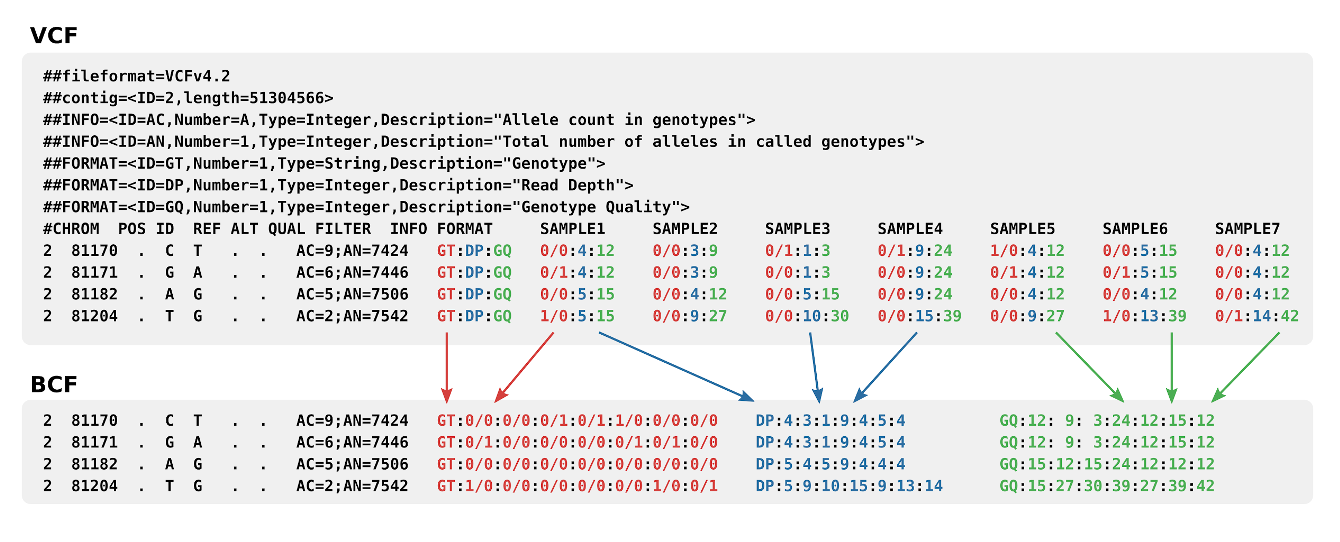
An explanation of the format in picture form
* {{Bioinformatics Biological sequence format
 The Variant Call Format (VCF) specifies the format of a text file used in
The Variant Call Format (VCF) specifies the format of a text file used in bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combin ...
for storing gene sequence variations. The format has been developed with the advent of large-scale genotyping
Genotyping is the process of determining differences in the genetic make-up (genotype) of an individual by examining the individual's DNA sequence using biological assays and comparing it to another individual's sequence or a reference sequence. ...
and DNA sequencing projects, such as the 1000 Genomes Project
The 1000 Genomes Project (abbreviated as 1KGP), launched in January 2008, was an international research effort to establish by far the most detailed catalogue of human genetic variation. Scientists planned to sequence the genomes of at least one th ...
. Existing formats for genetic data such as General feature format (GFF) stored all of the genetic data, much of which is redundant because it will be shared across the genomes. By using the variant call format only the variations need to be stored along with a reference genome.
The standard is currently in version 4.3, although the 1000 Genomes Project
The 1000 Genomes Project (abbreviated as 1KGP), launched in January 2008, was an international research effort to establish by far the most detailed catalogue of human genetic variation. Scientists planned to sequence the genomes of at least one th ...
has developed its own specification for structural variations such as duplications, which are not easily accommodated into the existing schema. There is also a genomic VCF (gVCF) extended format, which includes additional information about "blocks" that match the reference and their qualities. A set of tools is also available for editing and manipulating the files.
Example
##fileformat=VCFv4.3 ##fileDate=20090805 ##source=myImputationProgramV3.1 ##reference=file:///seq/references/1000GenomesPilot-NCBI36.fasta ##contig=The VCF header
The header begins the file and provides metadata describing the body of the file. Header lines are denoted as starting with . Special keywords in the header are denoted with . Recommended keywords include , and . The header contains keywords that optionally semantically and syntactically describe the fields used in the body of the file, notably INFO, FILTER, and FORMAT (see below).The columns of a VCF
The body of VCF follows the header, and is tab separated into 8 mandatory columns and an unlimited number of optional columns that may be used to record other information about the sample(s). When additional columns are used, the first optional column is used to describe the format of the data in the columns that follow.Common INFO fields
Arbitrary keys are permitted, although the following sub-fields are reserved (albeit optional): Any other info fields are defined in the .vcf header.Common FORMAT fields
Any other format fields are defined in the .vcf header.See also
* TheFASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics.
History
The original FASTA program ...
format, used to represent genome sequences.
* The FASTQ
FASTQ format is a text-based format for storing both a biological sequence (usually nucleotide sequence) and its corresponding quality scores. Both the sequence letter and quality score are each encoded with a single ASCII character for brevity.
...
format, used to represent DNA sequencer reads along with quality scores.
* The SAM
Sam, SAM or variants may refer to:
Places
* Sam, Benin
* Sam, Boulkiemdé, Burkina Faso
* Sam, Bourzanga, Burkina Faso
* Sam, Kongoussi, Burkina Faso
* Sam, Iran
* Sam, Teton County, Idaho, United States, a populated place
People and fictional c ...
format, used to represent genome sequencer reads that have been aligned to genome sequences.
* The GVF format (Genome Variation Format), an extension based on the GFF3 format.
* Global Alliance for Genomics and Health (GA4GH), the group leading the management and expansion of the VCF format. The VCF specification is no longer maintained by the 1000 Genomes Project.
* Human genome
The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the ...
* Human genetic variation
Human genetic variation is the genetic differences in and among populations. There may be multiple variants of any given gene in the human population (alleles), a situation called polymorphism.
No two humans are genetically identical. Even m ...
* Single Nucleotide Polymorphism (SNP)
References
External links
An explanation of the format in picture form
* {{Bioinformatics Biological sequence format