U6 SnRNA on:
[Wikipedia]
[Google]
[Amazon]
U6 snRNA is the
 The U6 snRNA is known to form an extensive base-pair interactions with
The U6 snRNA is known to form an extensive base-pair interactions with
 Free U6 snRNA is found to be associated with the proteins Prp24 and the
Free U6 snRNA is found to be associated with the proteins Prp24 and the
non-coding
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules (e.g. transfer RNA, microRNA, piRNA, ribosomal RNA, and regula ...
small nuclear RNA
Small nuclear RNA (snRNA) is a class of small RNA molecules that are found within the Cell nucleus#Splicing speckles, splicing speckles and Cajal body, Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approxi ...
(snRNA) component of U6 snRNP
snRNPs (pronounced "snurps"), or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre- ...
(''small nuclear ribonucleoprotein''), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA
A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by Transcription (genetics), transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcript ...
, and various other proteins to assemble a spliceosome
A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs ( snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to sp ...
, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA
A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by Transcription (genetics), transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcript ...
. Splicing, or the removal of introns
An intron is any Nucleic acid sequence, nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of ...
, is a major aspect of post-transcriptional modification
Transcriptional modification or co-transcriptional modification is a set of biological processes common to most eukaryotic cells by which an RNA primary transcript is chemically altered following transcription from a gene to produce a mature, f ...
and takes place only in the nucleus
Nucleus (: nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
*Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucleu ...
of eukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of ...
.
The RNA sequence of U6 is the most highly conserved across species of all five of the snRNAs involved in the spliceosome, suggesting that the function of the U6 snRNA has remained both crucial and unchanged through evolution.
It is common in vertebrate genomes to find many copies of the U6 snRNA gene or U6-derived pseudogenes. This prevalence of "back-ups" of the U6 snRNA gene in vertebrates further implies its evolutionary importance to organism viability.
The U6 snRNA gene has been isolated in many organisms, including '' C. elegans''. Among them, baker's yeast (''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungal microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have be ...
'') is a commonly used model organism
A model organism is a non-human species that is extensively studied to understand particular biological phenomena, with the expectation that discoveries made in the model organism will provide insight into the workings of other organisms. Mo ...
in the study of snRNAs.
The structure and catalytic mechanism of U6 snRNA resembles that of domain V of group II introns. The formation of the triple helix in U6 snRNA is deemed to be important in splicing activity, where its role is to bring the catalytic site to the splice site.
Role
Base-pair specificity of the U6 snRNA allows the U6 snRNP to bind tightly to the U4 snRNA and loosely to the U5 snRNA of a triple-snRNP during the initial phase of the splicing reaction. As the reaction progresses, the U6 snRNA is unzipped from U4 and binds to the U2 snRNA. At each stage of this reaction, the U6 snRNA secondary structure undergoes extensive conformational changes. The association of U6 snRNA with the 5' end of theintron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gen ...
via base-pairing during the splicing reaction occurs prior to the formation of the lariat
A lasso or lazo ( or ), also called reata or la reata in Mexico, and in the United States riata or lariat (from Mexican Spanish lasso for roping cattle), is a loop of rope designed as a restraint to be thrown around a target and tightened when ...
(or ''lasso-shaped'') intermediate, and is required for the splicing process to proceed. The association of U6 snRNP with U2 snRNP via base-pairing forms the U6-U2 complex, a structure that comprises the active site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the ''binding s ...
of the spliceosome
A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs ( snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to sp ...
.
Secondary structure
While the putative secondary structure consensus base pairing is confined to a short 5'stem-loop
Stem-loops are nucleic acid Biomolecular structure, secondary structural elements which form via intramolecular base pairing in single-stranded DNA or RNA. They are also referred to as hairpins or hairpin loops. A stem-loop occurs when two regi ...
, much more extensive structures have been proposed for specific organisms such as in yeast. In addition to the 5' stem loop, all confirmed U6 snRNAs can form the proposed 3' intramolecular stem loop.
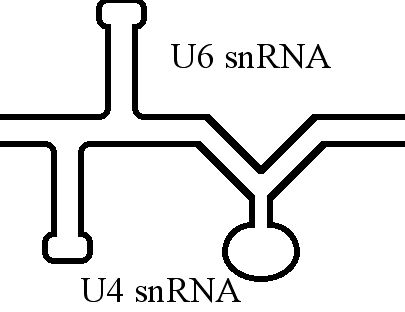
 The U6 snRNA is known to form an extensive base-pair interactions with
The U6 snRNA is known to form an extensive base-pair interactions with U4 snRNA U4, U-4, or U04 may refer to:
Science and technology
* U4 spliceosomal RNA, a non-coding RNA component of the major U2-dependent spliceosome
* Haplogroup U4 (mtDNA), a human genetic group
* U4, unitary group of degree 4
* U-47700, a synthetic opio ...
. This interaction has been shown to be mutually exclusive to that of the 3' intramolecular stem loop.
Associated proteins
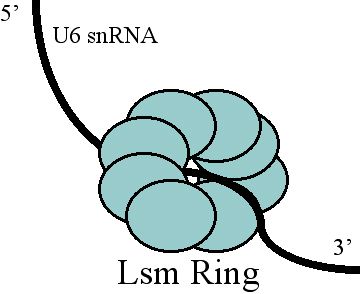
 Free U6 snRNA is found to be associated with the proteins Prp24 and the
Free U6 snRNA is found to be associated with the proteins Prp24 and the LSm
LSM may refer to:
Science
*Laboratoire Souterrain de Modane (Modane Underground Laboratory), a particle physics laboratory in France
*Lanthanum strontium manganite, a crystal used as a cathode material
*Confocal microscopy, Laser scanning microsc ...
s. Prp24 is thought to form an intermediate complex with the U6 snRNA, in order to facilitate the extensive base-pairing between the U4 and U6 snRNAs, and the Lsms may aid in Prp24 binding. The approximate location of these protein binding domains was determined, and the proteins were later visualized by electron microscopy. This study suggests that in the free form of U6, Prp24 binds to the telestem and the uridine-rich 3' tail of the U6 snRNA is threaded through the ring of Lsms. Another important NTC-related protein associated with U6 is Cwc2, which by interaction with important catalytic RNA elements induces the formation of a functional catalytic core in the spliceosome. Cwc2 and U6 achieve formation of this complex by interaction with the ISL and regions located near the 5' splice site.
See also
* U6atac minor spliceosomal RNAReferences
Further reading
*External links
* {{Rfam, id=RF00026, name=U6 spliceosomal RNA Small nuclear RNA Spliceosome RNA splicing