Tobacco Etch Virus Protease on:
[Wikipedia]
[Google]
[Amazon]
TEV protease (, ''Tobacco Etch Virus nuclear-inclusion-a endopeptidase'') is a highly sequence-specific
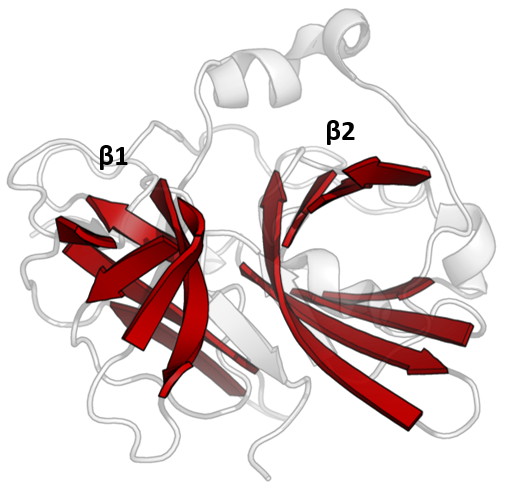 The structure of TEV protease has been solved by
The structure of TEV protease has been solved by
 The preferred, native cleavage sequence was first identified by examining the cut sites in the native polyprotein substrate for recurring sequence. The consensus for these native cut sites is ENLYFQ\S where ‘\’ denotes the cleaved peptide bond. Residues of the substrate are labelled P6 to P1 before the cut site and P1’ after the cut site. Early works also measured cleavage of an array of similar substrates to characterise how specific the protease was for the native sequence.
Studies have subsequently used sequencing of cleaved substrates from a pool of randomised sequences to determine preference patterns. Although ENLYFQ\S is the optimal sequence, the protease is active to a greater or lesser extent on a range of substrates (i.e. shows some substrate promiscuity). The highest cleavage is of sequences closest to the consensus EXLYΦQ\φ where X is any residue, Φ is any large or medium hydrophobe and φ is any small hydrophobic or polar residue. Although this sequence is the optimal, sequences with disfavoured residues at some positions can still be cleaved if the rest of the sequence is optimal.
Specificity is endowed by the large contact area between enzyme and substrate. Proteases such as
The preferred, native cleavage sequence was first identified by examining the cut sites in the native polyprotein substrate for recurring sequence. The consensus for these native cut sites is ENLYFQ\S where ‘\’ denotes the cleaved peptide bond. Residues of the substrate are labelled P6 to P1 before the cut site and P1’ after the cut site. Early works also measured cleavage of an array of similar substrates to characterise how specific the protease was for the native sequence.
Studies have subsequently used sequencing of cleaved substrates from a pool of randomised sequences to determine preference patterns. Although ENLYFQ\S is the optimal sequence, the protease is active to a greater or lesser extent on a range of substrates (i.e. shows some substrate promiscuity). The highest cleavage is of sequences closest to the consensus EXLYΦQ\φ where X is any residue, Φ is any large or medium hydrophobe and φ is any small hydrophobic or polar residue. Although this sequence is the optimal, sequences with disfavoured residues at some positions can still be cleaved if the rest of the sequence is optimal.
Specificity is endowed by the large contact area between enzyme and substrate. Proteases such as
TEV polyprotein on UniProt
TEV protease on MEROPS
National Cancer Institute TEV FAQ
{{Portal bar, Biology, border=no EC 3.4.22 Proteases
cysteine protease
Cysteine proteases, also known as thiol proteases, are hydrolase enzymes that degrade proteins. These proteases share a common catalytic mechanism that involves a nucleophilic cysteine thiol in a catalytic triad or dyad.
Discovered by Gopal Chu ...
from Tobacco Etch Virus (TEV). It is a member of the PA clan
The PA clan ( Proteases of mixed nucleophile, superfamily A) is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but can have iden ...
of chymotrypsin
Chymotrypsin (, chymotrypsins A and B, alpha-chymar ophth, avazyme, chymar, chymotest, enzeon, quimar, quimotrase, alpha-chymar, alpha-chymotrypsin A, alpha-chymotrypsin) is a digestive enzyme component of pancreatic juice acting in the duodenu ...
-like proteases. Due to its high sequence specificity, TEV protease is frequently used for the controlled cleavage of fusion proteins ''in vitro
''In vitro'' (meaning ''in glass'', or ''in the glass'') Research, studies are performed with Cell (biology), cells or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in ...
'' and ''in vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, an ...
''. The consensus sequence recognized by TEV protease is Glu-Asn-Leu-Tyr-Phe-Gln-, -Ser, where ", " denotes cleaved peptide bond.
Origin
The tobacco etch virus encodes its entire genome as a single massivepolyprotein
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Protein degradation is a major regulatory mechanism of gene expression and contributes substantially to shaping mammalian proteomes. Uncatalysed, the hydrolysis o ...
(350 kDa). This is cleaved into functional units by the three proteases: P1 protease (1 cleavage site), helper-component protease (1 cleavage site) and TEV protease (7 cleavage sites).UniProt: TEV polyprotein: The native TEV protease also contains an internal self-cleavage site. This site is slowly cleaved to inactivate the enzyme (the physiological reason for this is unknown).
Structure and function
 The structure of TEV protease has been solved by
The structure of TEV protease has been solved by X-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
. It is composed of two β-barrels and a flexible C-terminal tail and displays structural homology to the chymotrypsin superfamily of proteases (PA clan
The PA clan ( Proteases of mixed nucleophile, superfamily A) is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but can have iden ...
, C4 family by MEROPS
MEROPS is an online database for peptidases (also known as proteases, proteinases and proteolytic enzymes) and their inhibitors. The classification scheme for peptidases was published by Rawlings & Barrett in 1993, and that for protein inhibito ...
classification). Although homologous to cellular serine proteases
Serine proteases (or serine endopeptidases) are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site.
They are found ubiquitously in both eukaryotes and prokaryotes. Seri ...
(such as trypsin
Trypsin is an enzyme in the first section of the small intestine that starts the digestion of protein molecules by cutting long chains of amino acids into smaller pieces. It is a serine protease from the PA clan superfamily, found in the dig ...
, elastase
In molecular biology, elastase is an enzyme from the class of proteases (peptidases) that break down proteins, specifically one that can break down elastin. In other words, the name only refers to the substrate specificity (i.e. what proteins i ...
, thrombin
Prothrombin (coagulation factor II) is encoded in the human by the F2-gene. It is proteolytically cleaved during the clotting process by the prothrombinase enzyme complex to form thrombin.
Thrombin (Factor IIa) (, fibrose, thrombase, throm ...
etc.), TEV protease uses a cysteine as its catalytic nucleophile (as do many other viral proteases).
Covalent catalysis is performed with an Asp-His-Cys triad, split between the two barrels (Asp on β1 and His and Cys on β2). The substrate is held as a β-sheet, forming an antiparallel interaction with the cleft between the barrels and a parallel interaction with the C-terminal tail. The enzyme therefore forms a binding tunnel around the substrate and side chain interactions control specificity.
Specificity
 The preferred, native cleavage sequence was first identified by examining the cut sites in the native polyprotein substrate for recurring sequence. The consensus for these native cut sites is ENLYFQ\S where ‘\’ denotes the cleaved peptide bond. Residues of the substrate are labelled P6 to P1 before the cut site and P1’ after the cut site. Early works also measured cleavage of an array of similar substrates to characterise how specific the protease was for the native sequence.
Studies have subsequently used sequencing of cleaved substrates from a pool of randomised sequences to determine preference patterns. Although ENLYFQ\S is the optimal sequence, the protease is active to a greater or lesser extent on a range of substrates (i.e. shows some substrate promiscuity). The highest cleavage is of sequences closest to the consensus EXLYΦQ\φ where X is any residue, Φ is any large or medium hydrophobe and φ is any small hydrophobic or polar residue. Although this sequence is the optimal, sequences with disfavoured residues at some positions can still be cleaved if the rest of the sequence is optimal.
Specificity is endowed by the large contact area between enzyme and substrate. Proteases such as
The preferred, native cleavage sequence was first identified by examining the cut sites in the native polyprotein substrate for recurring sequence. The consensus for these native cut sites is ENLYFQ\S where ‘\’ denotes the cleaved peptide bond. Residues of the substrate are labelled P6 to P1 before the cut site and P1’ after the cut site. Early works also measured cleavage of an array of similar substrates to characterise how specific the protease was for the native sequence.
Studies have subsequently used sequencing of cleaved substrates from a pool of randomised sequences to determine preference patterns. Although ENLYFQ\S is the optimal sequence, the protease is active to a greater or lesser extent on a range of substrates (i.e. shows some substrate promiscuity). The highest cleavage is of sequences closest to the consensus EXLYΦQ\φ where X is any residue, Φ is any large or medium hydrophobe and φ is any small hydrophobic or polar residue. Although this sequence is the optimal, sequences with disfavoured residues at some positions can still be cleaved if the rest of the sequence is optimal.
Specificity is endowed by the large contact area between enzyme and substrate. Proteases such as trypsin
Trypsin is an enzyme in the first section of the small intestine that starts the digestion of protein molecules by cutting long chains of amino acids into smaller pieces. It is a serine protease from the PA clan superfamily, found in the dig ...
have specificity for one residue before and after the cleaved bond due to a shallow binding cleft with only one or two pockets that bind the substrate side chains. Conversely, viral proteases such as TEV protease have a long C-terminal tail which completely covers the substrate to create a binding tunnel. This tunnel contains a set of tight binding pockets such that each side chain of the substrate peptide (P6 to P1’) is bound in a complementary site (S6 to S1’).
In particular, peptide side chain P6-Glu contacts a network of three hydrogen bonds; P5-Asn points into the solvent, making no specific interactions (hence the absence of substrate consensus at this position); P4-Leu is buried in a hydrophobic pocket; P3-Tyr is held in a hydrophobic pocket with a short hydrogen bond at the end; P2-Phe is also surrounded by hydrophobes including the face of the triad histidine; P1-Gln forms four hydrogen bonds; and P1’-Ser is only partly enclosed in a shallow hydrophobic groove.
Application as a biochemical tool
One of the main uses of this protein is for removing affinity tags from purified recombinant fusion proteins. The reason for the use of TEV protease as a biochemical tool is its high sequence specificity. This specificity allows for the controlled cleavage of proteins when the preference sequence is inserted into flexible loops. It also makes TEV protease relatively non-toxic ''in vivo'' as the recognized sequence scarcely occurs in proteins. Although rational design has had limited success in changing protease specificity,directed evolution
Directed evolution (DE) is a method used in protein engineering that mimics the process of natural selection to steer proteins or nucleic acids toward a user-defined goal. It consists of subjecting a gene to iterative rounds of mutagenesis (cre ...
has been used to change the preferred residue either before or after the cleavage site.
However, TEV protease does have limitations as a biochemical tool. It is prone to deactivation by self-cleavage (autolysis), though this can be abolished through a single S219V mutation in the internal cleavage site. The protease expressed alone is also poorly soluble, however several attempts have been made to improve its solubility through directed evolution
Directed evolution (DE) is a method used in protein engineering that mimics the process of natural selection to steer proteins or nucleic acids toward a user-defined goal. It consists of subjecting a gene to iterative rounds of mutagenesis (cre ...
and computational design. It has also been shown that expression can be improved by fusion to maltose binding protein
Maltose-binding protein (MBP) is a part of the maltose/maltodextrin system of ''Escherichia coli'', which is responsible for the uptake and efficient catabolism of maltodextrins. It is a complex regulatory and transport system involving many prote ...
(MBP) which acts a solubility-enhancing partner.
TEV protease has been reported to show a 10-fold loss of activity at 4 °C. TEV protease shows loss of activity at temperatures above 34 °C. The original TEV protease required the presence of reducing agent for high activity, which could interfere with the function of proteins containing disulfide bonds. After incorporation of various mutations, later "superTEV protease" versions are highly active in the presence or absence of reducing agent.
The molecular weight of this enzyme varies between 25 and 27 kDa depending on the specific construct used.
References
External links
TEV polyprotein on UniProt
TEV protease on MEROPS
National Cancer Institute TEV FAQ
{{Portal bar, Biology, border=no EC 3.4.22 Proteases