Structural Variant on:
[Wikipedia]
[Google]
[Amazon]
Genomic structural variation is the variation in structure of an organism's
 New methods have been developed to analyze human genetic structural variation at high resolutions. The methods used to test the genome are in either a specific targeted way or in a genome wide manner. For Genome wide tests, array-based comparative genome hybridization approaches bring the best genome wide scans to find new copy number variants. These techniques use DNA fragments that are labeled from a genome of interest and are hybridized, with another genome labeled differently, to arrays spotted with cloned DNA fragments. This reveals copy number differences between two genomes.
For targeted genome examinations, the best assays for checking specific areas of the genome are primarily PCR based. The best established of the PCR based methods is real time quantitative polymerase chain reaction (qPCR). A different approach is to specifically check certain areas that surround known segmental duplications since they are usually areas of copy number variation. An SNP genotyping method that offers independent fluorescence intensities for two alleles can be used to target the nucleotides in between two copies of a segmental duplication. From this, an increase in intensity from one of the alleles compared to the other can be observed.
With the development of
New methods have been developed to analyze human genetic structural variation at high resolutions. The methods used to test the genome are in either a specific targeted way or in a genome wide manner. For Genome wide tests, array-based comparative genome hybridization approaches bring the best genome wide scans to find new copy number variants. These techniques use DNA fragments that are labeled from a genome of interest and are hybridized, with another genome labeled differently, to arrays spotted with cloned DNA fragments. This reveals copy number differences between two genomes.
For targeted genome examinations, the best assays for checking specific areas of the genome are primarily PCR based. The best established of the PCR based methods is real time quantitative polymerase chain reaction (qPCR). A different approach is to specifically check certain areas that surround known segmental duplications since they are usually areas of copy number variation. An SNP genotyping method that offers independent fluorescence intensities for two alleles can be used to target the nucleotides in between two copies of a segmental duplication. From this, an increase in intensity from one of the alleles compared to the other can be observed.
With the development of
The 1000 Genomes Project
Congenital disorders Chromosomes
chromosome
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most import ...
, such as deletions, duplications, copy-number variants
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of G ...
, insertions, inversions and translocations
In genetics, chromosome translocation is a phenomenon that results in unusual rearrangement of chromosomes. This includes "balanced" and "unbalanced" translocation, with three main types: "reciprocal", "nonreciprocal" and "Robertsonian" transloc ...
. Originally, a structure variation affects a sequence length about 1kb to 3Mb, which is larger than SNPs
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in ...
and smaller than chromosome abnormality
A chromosomal abnormality, chromosomal anomaly, chromosomal aberration, chromosomal mutation, or chromosomal disorder is a missing, extra, or irregular portion of chromosomal DNA. These can occur in the form of numerical abnormalities, where the ...
(though the definitions have some overlap). However, the operational range of structural variants has widened to include events > 50bp. Some structural variants are associated with genetic diseases
A genetic disorder is a health problem caused by one or more abnormalities in the genome. It can be caused by a mutation in a single gene (monogenic) or multiple genes (polygenic) or by a chromosome abnormality. Although polygenic disorders are ...
, however most are not. Approximately 13% of the human genome is defined as structurally variant in the normal population, and there are at least 240 genes that exist as homozygous
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism.
Mos ...
deletion polymorphisms in human populations, suggesting these genes are dispensable in humans. While humans carry a median of 3.6 Mbp in SNPs (compared to a reference genome), a median of 8.9 Mbp is affected by structural variation which thus causes most genetic differences between humans in terms of raw sequence data.
Microscopic structural variation
Microscopic means that it can be detected withoptical microscope
The optical microscope, also referred to as a light microscope, is a type of microscope that commonly uses visible light and a system of lenses to generate magnified images of small objects. Optical microscopes are the oldest design of micros ...
s, such as aneuploidies, marker chromosome
A marker chromosome (mar) is a small fragment of a chromosome which generally cannot be identified without specialized genomic analysis due to the size of the fragment.Thompson & Thompson Genetics in Medicine, Chapter 5, 57-74 https://www.clinicalk ...
, gross rearrangements and variation in chromosome size. The frequency in human population is thought to be underestimated due to the fact that some of these are not actually easy to identify. These structural abnormalities exist in 1 of every 375 live births by putative information.
Sub-microscopic structural variation
Sub-microscopic structural variants are much harder to detect owing to their small size. The first study in 2004 that usedDNA microarray
A DNA microarray (also commonly known as a DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or t ...
s could detect tens of genetic loci that exhibited copy number variation
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of ...
, deletions and duplications, greater than 100 kilobase
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s in the human genome. However, by 2015 whole genome sequencing studies could detect around 5,000 of structural variants as small as 100 base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s encompassing approximately 20 megabase
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s in each individual genome. These structural variants include deletions, tandem duplications, inversions, mobile element insertions. The mutation rate is also much higher than microscopic structural variants, estimated by two studies at 16% and 20% respectively, both of which are probably underestimates due to the challenges of accurately detecting structural variants. It has also been shown that the generation of spontaneous structural variants significantly increases the likelihood of generating further spontaneous single nucleotide variants or indel
Indel (insertion-deletion) is a molecular biology term for an insertion or deletion of bases in the genome of an organism. Indels ≥ 50 bases in length are classified as structural variants.
In coding regions of the genome, unless the lengt ...
s within 100 kilobase
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s of the structural variation event.
Copy-number variation
Copy-number variation (CNV) is a large category of structural variation, which includes insertions, deletions and duplications. In recent studies, copy-number variations are tested on people who do not have genetic diseases, using methods that are used for quantitative SNP genotyping. Results show that 28% of the suspected regions in the individuals actually do contain copy number variations. Also, CNVs in human genome affect more nucleotides thanSingle Nucleotide Polymorphism
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in ...
(SNP).
It is also noteworthy that many of CNVs are not in coding regions. Because CNVs are usually caused by unequal recombination, widespread similar sequences such as LINEs and SINE
In mathematics, sine and cosine are trigonometric functions of an angle. The sine and cosine of an acute angle are defined in the context of a right triangle: for the specified angle, its sine is the ratio of the length of the side opposite th ...
s may be a common mechanism of CNV creation.
Inversion
There are several inversions known which are related to human disease. For instance, recurrent 400kb inversion in factor VIII gene is a common cause ofhaemophilia A
Haemophilia A (or hemophilia A) is a blood clotting disorder caused by a genetic deficiency in clotting factor VIII, thereby resulting in significant susceptibility to bleeding, both internally and externally. This condition occurs almost exclu ...
, and smaller inversions affecting idunorate 2-sulphatase (IDS) will cause Hunter syndrome
Hunter syndrome, or mucopolysaccharidosis type II (MPS II), is a rare genetic disorder, inherited lysosomal storage disease in which large sugar molecules called glycosaminoglycans (or GAGs or mucopolysaccharides) build up in body tissues. Hunte ...
. More examples include Angelman syndrome
Angelman syndrome (AS) is a genetic disorder that affects approximately 1 in 15,000 individuals. AS impairs the function of the nervous system, producing symptoms, such as severe intellectual disability, developmental disability, limited to no ...
and Sotos syndrome
Sotos syndrome is a rare genetic disorder characterized by excessive physical growth during the first years of life. Excessive growth often starts in infancy and continues into the early teen years. The disorder may be accompanied by autism, mild ...
. However, recent research shows that one person can have 56 putative inversions, thus the non-disease inversions are more common than previously supposed. Also in this study it's indicated that inversion breakpoints are commonly associated with segmental duplications. One 900 kb inversion in the chromosome 17
Chromosome 17 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 17 spans more than 84 million base pairs (the building material of DNA) and represents between 2.5 and 3% of the total DN ...
is under positive selection
In population genetics, directional selection is a type of natural selection in which one extreme phenotype is favored over both the other extreme and moderate phenotypes. This genetic selection causes the allele frequency to shift toward the ...
and are predicted to increase its frequency in European population.
Other structural variants
More complex structural variants can occur include a combination of the above in a single event. The most common type of complex structural variation are non-tandem duplications, where sequence is duplicated and inserted in inverted or direct orientation into another part of the genome. Other classes of complex structural variant include deletion-inversion-deletions, duplication-inversion-duplications, and tandem duplications with nested deletions. There are also cryptic translocations and segmental uniparental disomy (UPD). There are increasing reports of these variations, but are more difficult to detect than traditional variations because these variants are balanced and array-based or PCR-based methods are not able to locate them.Structural variation and phenotypes
Somegenetic diseases
A genetic disorder is a health problem caused by one or more abnormalities in the genome. It can be caused by a mutation in a single gene (monogenic) or multiple genes (polygenic) or by a chromosome abnormality. Although polygenic disorders are ...
are suspected to be caused by structural variations, but the relation is not very certain. It is not plausible to divide these variants into two classes as "normal" or "disease", because the actual output of the same variant will also vary. Also, a few of the variants are actually positively selected for (mentioned above).
A series of studies have shown that gene disrupting spontaneous (''de novo'') CNVs disrupt genes approximately four times more frequently in autism than in controls and contribute to approximately 5–10% of cases. Inherited variants also contribute to around 5–10% of cases of autism.
Structural variations also have its function in population genetics. Different frequency of a same variation can be used as a genetic mark to infer relationship between populations in different areas. A complete comparison between human and chimpanzee structural variation also suggested that some of these may be fixed in one species because of its adaptative function. There are also deletions related to resistance against malaria
Malaria is a Mosquito-borne disease, mosquito-borne infectious disease that affects vertebrates and ''Anopheles'' mosquitoes. Human malaria causes Signs and symptoms, symptoms that typically include fever, Fatigue (medical), fatigue, vomitin ...
and AIDS
The HIV, human immunodeficiency virus (HIV) is a retrovirus that attacks the immune system. Without treatment, it can lead to a spectrum of conditions including acquired immunodeficiency syndrome (AIDS). It is a Preventive healthcare, pr ...
. Also, some highly variable segments are thought to be caused by balancing selection, but there are also studies against this hypothesis.
Database of structural variation
Some of genome browsers andbioinformatic
Bioinformatics () is an interdisciplinary field of science
Science is a systematic discipline that builds and organises knowledge in the form of testable hypotheses and predictions about the universe. Modern science is typically divi ...
databases have a list of structural variations in human genome with an emphasis on CNVs, and can show them in the genome browsing page, for example, UCSC Genome Browser
The UCSC Genome Browser is an online and downloadable genome browser hosted by the University of California, Santa Cruz (UCSC). It is an interactive website offering access to genome sequence data from a variety of vertebrate and invertebrate spec ...
. Under the page viewing a part of the genome, there are "Common Cell CNVs" and "Structural Var" which can be enabled.
On NCBI, there is a special page for structural variation. In that system, both "inner" and "outer" coordinates are shown; they are both not actual breakpoints, but surmised minimal and maximum range of sequence affected by the structural variation. The types are classified as insertion, loss, gain, inversion, LOH, everted, transchr and UPD.
Methods of detection
 New methods have been developed to analyze human genetic structural variation at high resolutions. The methods used to test the genome are in either a specific targeted way or in a genome wide manner. For Genome wide tests, array-based comparative genome hybridization approaches bring the best genome wide scans to find new copy number variants. These techniques use DNA fragments that are labeled from a genome of interest and are hybridized, with another genome labeled differently, to arrays spotted with cloned DNA fragments. This reveals copy number differences between two genomes.
For targeted genome examinations, the best assays for checking specific areas of the genome are primarily PCR based. The best established of the PCR based methods is real time quantitative polymerase chain reaction (qPCR). A different approach is to specifically check certain areas that surround known segmental duplications since they are usually areas of copy number variation. An SNP genotyping method that offers independent fluorescence intensities for two alleles can be used to target the nucleotides in between two copies of a segmental duplication. From this, an increase in intensity from one of the alleles compared to the other can be observed.
With the development of
New methods have been developed to analyze human genetic structural variation at high resolutions. The methods used to test the genome are in either a specific targeted way or in a genome wide manner. For Genome wide tests, array-based comparative genome hybridization approaches bring the best genome wide scans to find new copy number variants. These techniques use DNA fragments that are labeled from a genome of interest and are hybridized, with another genome labeled differently, to arrays spotted with cloned DNA fragments. This reveals copy number differences between two genomes.
For targeted genome examinations, the best assays for checking specific areas of the genome are primarily PCR based. The best established of the PCR based methods is real time quantitative polymerase chain reaction (qPCR). A different approach is to specifically check certain areas that surround known segmental duplications since they are usually areas of copy number variation. An SNP genotyping method that offers independent fluorescence intensities for two alleles can be used to target the nucleotides in between two copies of a segmental duplication. From this, an increase in intensity from one of the alleles compared to the other can be observed.
With the development of next-generation sequencing
Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of massively parallel processing; it is also called next-generation sequencing (NGS) or second-generation ...
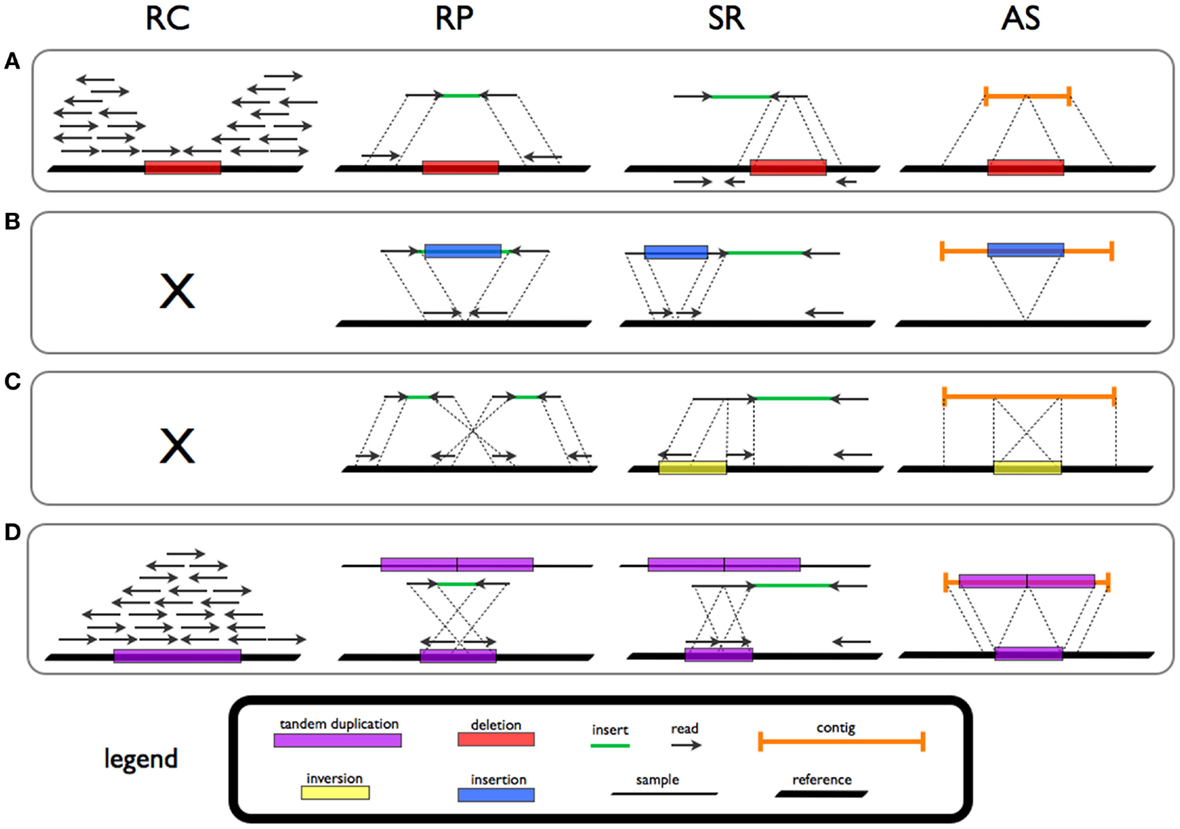
(NGS) technology, four classes of strategies for the detection of structural variants with NGS data have been reported, with each being based on patterns that are diagnostic of different classes of SV.
* Read-depth or read-count methods assume a random distribution (e.g. Poisson distribution
In probability theory and statistics, the Poisson distribution () is a discrete probability distribution that expresses the probability of a given number of events occurring in a fixed interval of time if these events occur with a known const ...
) of reads from short read sequencing. The divergence from this distribution is investigated to discover duplications and deletions. Regions with duplication will show higher read depth while those with deletion will result in lower read depth.
* Split-read methods enable detection of insertions (including mobile element insertions) and deletions down to single base-pair resolution. The presence of a SV is identified from discontinuous alignment to the reference genome. A gap in the read marks a deletion and in the reference marks an insertion.
* Read pair methods examine the length and orientation of paired-end reads from short read sequencing data. For example, read pairs further apart than expected indicate a deletion. Translocations, inversions and tandem duplications can likewise be discovered using read-pairs.
* ''De novo'' sequence assembly may be applied with reads that are accurate enough. While, in practice, use of this method is limited by the length of sequence reads, long read based genome assemblies offer structural variation discovery for classes such as insertions that escape detection when using other methods.
See also
* Structural variation in the human genomeReferences
{{ReflistExternal links
The 1000 Genomes Project
Congenital disorders Chromosomes