Short Linear Motif on:
[Wikipedia]
[Google]
[Amazon]
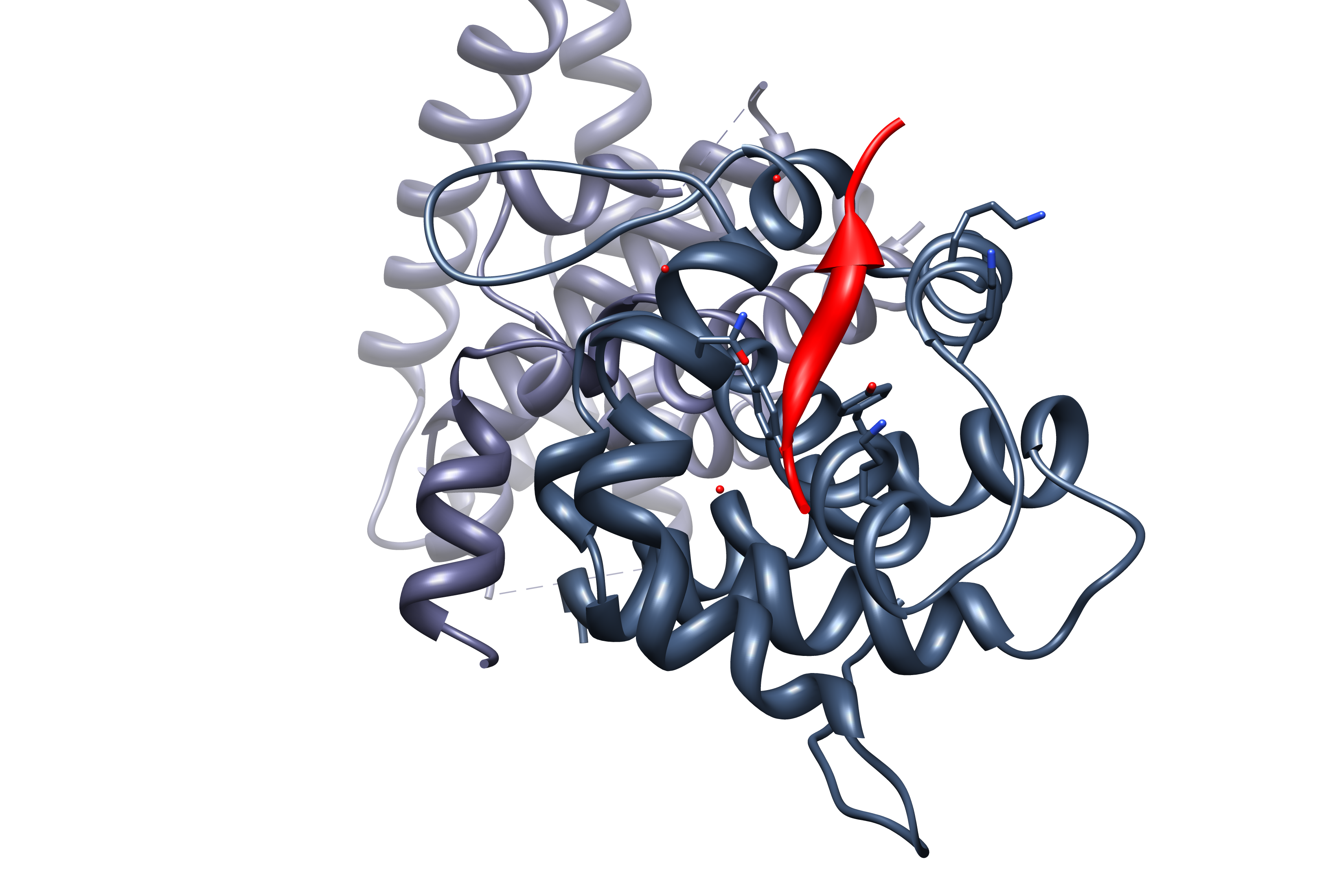 In molecular biology short linear motifs (SLiMs), linear motifs or minimotifs are short stretches of
In molecular biology short linear motifs (SLiMs), linear motifs or minimotifs are short stretches of
AP2-binding motifs
i
Eps15
.
Modification sites SLiMs encompass sites with intrinsic specificity determinant that are recognised and modified by the active site of a catalytic domain of an enzyme. These SLiMs include many classical post translational modification sites (PTMs), proteolytic cleavage sites recognised by proteases and bonds recognised by isomerases. * ''Moiety addition'' – SLiMs are often targeted for the addition of a small chemical groups (e.g.
Ligand binding site SLiMs recruit binding partners to the SLiM containing proteins, often mediating transient interactions, or acting co-operatively to produce more stable complexes. Ligand SLiMs are often central to the formation of dynamic multi-protein complexes, however, they more commonly mediate regulatory interactions that control the stability, localisation or modification state of a protein. * ''Complex formation'' – Ligand SLiMs often function as simple interfaces that recruit proteins to multi-protein complexes (e.g. the Retinoblastoma-binding LxCxE motif) or act as aggregators in scaffold proteins (e.g.

Pawsons Lab Resource on motif-binding domains
Eukaryotic Linear Motif Database
MiniMotif Miner
PepCyber
ScanSite
ANCHOR
DiLiMot
Eukaryotic Linear Motif Database
MiniMotif Miner
SLiMSuite
: *
SLiMPred
*
*
*
ScanSite
{{MotifBindingDomains Protein structural motifs Protein domains
 In molecular biology short linear motifs (SLiMs), linear motifs or minimotifs are short stretches of
In molecular biology short linear motifs (SLiMs), linear motifs or minimotifs are short stretches of protein sequence
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthe ...
that mediate protein–protein interaction
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and t ...
.
The first definition was given by Tim Hunt
Sir Richard Timothy Hunt (born 19 February 1943) is a British biochemist and molecular physiologist. He was awarded the 2001 Nobel Prize in Physiology or Medicine with Paul Nurse and Leland H. Hartwell for their discoveries of protein molecu ...
:
"The sequences of many proteins contain short, conserved motifs that are involved in recognition and targeting activities, often separate from other functional properties of the molecule in which they occur. These motifs are linear, in the sense that three-dimensional organization is not required to bring distant segments of the molecule together to make the recognizable unit. The conservation of these motifs varies: some are highly conserved while others, for example, allow substitutions that retain only a certain pattern of charge across the motif."
Attributes
SLiMs are generally situated in intrinsically disordered regions (over 80% of known SLiMs), however, upon interaction with a structured partnersecondary structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta ...
is often induced. The majority of annotated SLiMs consist of 3 to 11 contiguous amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
s, with an average of just over 6 residues. However, only few hotspot residues (on average 1 hotspot for each 3 residues in the motif) contribute the majority of the free energy of binding and determine most of the affinity and specificity of the interaction. Although most motifs have no positional preference, several of them are required to be localized at the protein termini in order to be functional.
The key defining attribute of SLiMs, having a limited number of residues that directly contact the binding partner, has two major consequences. First, only few or even a single mutation can result in the generation of a functional motif, with further mutations of flanking residues allowing tuning affinity and specificity. This results in SLiMs having an increased propensity to evolve convergently, which facilitates their proliferation, as is evidenced by their conservation and increased incidence in higher Eukaryote
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
s. It has been hypothesized that this might increase and restructure the connectivity of the interactome
In molecular biology, an interactome is the whole set of molecular interactions in a particular cell. The term specifically refers to physical interactions among molecules (such as those among proteins, also known as protein–protein interactions ...
. Second, SLiMs have relatively low affinity for their interaction partners (generally between 1 and 150 μM), which makes these interactions transient and reversible, and thus ideal to mediate dynamic processes such as cell signaling
In biology, cell signaling (cell signalling in British English) is the Biological process, process by which a Cell (biology), cell interacts with itself, other cells, and the environment. Cell signaling is a fundamental property of all Cell (biol ...
. In addition, this means that these interactions can be easily modulated by post-translational modifications
In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translation (biolog ...
that change the structural and physicochemical properties of the motif. Also, regions of high functional density can mediate molecular switch
A molecular switch is a molecule that can be switched between two or more stable or Metastability, metastable states with the use of any external (exogenous) or internal (endogenous) stimuli, such as changes in pH, light, temperature, an electri ...
ing by means of overlapping motifs (e.g. the C-terminal tails of integrin
Integrins are transmembrane receptors that help cell–cell and cell–extracellular matrix (ECM) adhesion. Upon ligand binding, integrins activate signal transduction pathways that mediate cellular signals such as regulation of the cell cycle, o ...
beta subunits), or they can allow high avidity
In biochemistry, avidity refers to the accumulated strength of ''multiple'' affinities of individual non-covalent binding interactions, such as between a protein receptor and its ligand, and is commonly referred to as functional affinity. Avidity ...
interactions by multiple low affinity motifs (e.g. multiplAP2-binding motifs
i
Eps15
.
Function
SLiM functions in almost every pathway due to their critical role in regulatory function, protein-protein interaction and signal transduction. SLiM act as interaction modules that are recognised by additional biomolecules. The majority of known interaction partners of SLiMs are globular protein domains, though, SLiMs that recognise other intrinsically disordered regions, RNA and lipids have also been characterised. SLiMs can be broadly split into two high level classes, modification sites and ligand binding sites. Modification sitesModification sites SLiMs encompass sites with intrinsic specificity determinant that are recognised and modified by the active site of a catalytic domain of an enzyme. These SLiMs include many classical post translational modification sites (PTMs), proteolytic cleavage sites recognised by proteases and bonds recognised by isomerases. * ''Moiety addition'' – SLiMs are often targeted for the addition of a small chemical groups (e.g.
Phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
), proteins (e.g. SUMOylation
In molecular biology, SUMO (Small Ubiquitin-like Modifier) proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation (pronounced ...
) or other moieties (e.g. post translational moiety addition).
* ''Proteolytic cleavage'' -SLiMs can act as recognition sites of endo-peptidases resulting in the irreversible cleavage of the peptide at the SLiM.
* ''Structural modifications'' – SLiMs can be recognised by isomerases resulting in the cis-trans isomerisation of the peptide backbone.
Ligand binding sitesLigand binding site SLiMs recruit binding partners to the SLiM containing proteins, often mediating transient interactions, or acting co-operatively to produce more stable complexes. Ligand SLiMs are often central to the formation of dynamic multi-protein complexes, however, they more commonly mediate regulatory interactions that control the stability, localisation or modification state of a protein. * ''Complex formation'' – Ligand SLiMs often function as simple interfaces that recruit proteins to multi-protein complexes (e.g. the Retinoblastoma-binding LxCxE motif) or act as aggregators in scaffold proteins (e.g.
SH3 domain
The SRC Homology 3 Domain (or SH3 domain) is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of ph ...
-binding proline-rich sequences).
* ''Localisation'' – A large number of SLiMs act as zipcodes that are recognized by the cellular transport machinery mediating the relocalisation of the containing protein to the correct sub-cellular compartment (e.g. Nuclear localisation signals (NLSs) and Nuclear export signal
A nuclear export signal (NES) is a short target peptide containing 4 hydrophobic residues in a protein that targets it for export from the cell nucleus to the cytoplasm through the nuclear pore complex using nuclear transport. It has the opposit ...
s (NESs))
* ''Modification state'' – Many classes of ligand SLiMs recruit enzymes to their substrate by binding to sites that are distinct from the enzyme's active site. These site, known as docking motifs, act as additional specificity determinants for these enzymes and decrease the likelihood of off-target modification events.
* ''Stability'' – A subset of docking motifs recruit E3 ubiquitin ligase to their substrates. The resulting polyubiquitination targets the substrate for proteosomal destruction.
Role in disease
Disordered protein elements like SLiMs are frequently found in factors that regulate gene expression. As a result, several diseases have been linked to mutations that alter key SLiM-mediated functions. For instance, one cause ofNoonan Syndrome
Noonan syndrome (NS) is a genetic disorder that may present with mildly unusual facial features, short height, congenital heart disease, bleeding problems, and skeletal malformations. Facial features include widely spaced eyes, light-colored ...
is a mutation in the protein Raf-1 which abrogates the interaction with 14-3-3 proteins mediated by corresponding short linear motifs and thereby deregulate the Raf-1 kinase
In biochemistry, a kinase () is an enzyme that catalyzes the transfer of phosphate groups from high-energy, phosphate-donating molecules to specific substrates. This process is known as phosphorylation, where the high-energy ATP molecule don ...
activity. Usher's Syndrome is the most frequent cause of hereditary deaf-blindness in humans and can be caused by mutations in either PDZ domain
The PDZ domain is a common structural domain of 80-90 Amino acid, amino-acids found in the Signal transduction, signaling proteins of bacteria, yeast, plants, viruses and animals. Proteins containing PDZ domains play a key role in anchoring recept ...
s in Harmonin or the corresponding PDZ interaction motifs in the SANS protein. Finally, Liddle's Syndrome
Liddle's syndrome, also called Liddle syndrome, is a genetic disorder inherited in an autosomal dominant manner that is characterized by early, and frequently severe, high blood pressure associated with low plasma renin activity, metabolic alkalo ...
has been implicated with autosomal dominant activating mutations in the WW interaction motif in the β-(SCNNB_HUMA) and γ-(SCNNG_HUMA) subunits of the Epithelial sodium channel ENaC. These mutations abrogate the binding to the ubiquitin ligase NEDD4
E3 ubiquitin-protein ligase NEDD4, also known as neural precursor cell expressed developmentally down-regulated protein 4 (whence "NEDD4") is an enzyme that is, in humans, encoded by the ''NEDD4'' gene.
NEDD4 is an E3 ubiquitin ligase enzyme, tha ...
, thereby inhibiting channel degradation and prolonging the half-life of ENaC
The epithelial sodium channel (ENaC), (also known as amiloride-sensitive sodium channel) is a membrane-bound ion channel that is selectively permeable to sodium ions (). It is assembled as a heterotrimer composed of three homologous subunits α ...
, ultimately resulting in increased Na+ reabsorption, plasma volume extension and hypertension.
Viruses often mimic human SLiMs to hijack and disrupt a host's cellular machinery, thereby adding functionality to their compact genomes without necessitating new virally encoded proteins. In fact, many motifs were originally discovered in viruses, such as the Retinoblastoma binding LxCxE motif and the UEV domain binding PTAP late domain. The short generation times and high mutation rates of viruses, in association with natural selection, has led to multiple examples of mimicry of host SLiMs in every step of the viral life cycle (Src binding motif PxxP in Nef modulates replication, WW domain binding PPxY mediates budding in Ebola virus, A Dynein Light Chain binding motif in Rabies virus is vital for host infection). The YGL motif (Tyrosine
-Tyrosine or tyrosine (symbol Tyr or Y) or 4-hydroxyphenylalanine is one of the 20 standard amino acids that are used by cells to synthesize proteins. It is a conditionally essential amino acid with a polar side group. The word "tyrosine" is ...
-Glycine
Glycine (symbol Gly or G; ) is an amino acid that has a single hydrogen atom as its side chain. It is the simplest stable amino acid. Glycine is one of the proteinogenic amino acids. It is encoded by all the codons starting with GG (G ...
-Leucine
Leucine (symbol Leu or L) is an essential amino acid that is used in the biosynthesis of proteins. Leucine is an α-amino acid, meaning it contains an α-amino group (which is in the protonated −NH3+ form under biological conditions), an α-Car ...
) is an integrin
Integrins are transmembrane receptors that help cell–cell and cell–extracellular matrix (ECM) adhesion. Upon ligand binding, integrins activate signal transduction pathways that mediate cellular signals such as regulation of the cell cycle, o ...
-binding motif present in several viral glycoprotein
The term viral protein refers to both the products of the genome of a virus and any host proteins incorporated into the viral particle. Viral proteins are grouped according to their functions, and groups of viral proteins include structural protein ...
s including Equine Herpes Virus (EHV) 1, EHV-4, and in rotavirus VP4. The extent of human SLiM mimicry is surprising with many viral proteins containing several functional SLiMs, for example, the Adenovirus protein E1A.
Pathogenic bacteria also mimic host motifs (as well as having their own motifs), however, not to the same extent as the obligate parasite viruses. E. Coli injects a protein, EspF(U), that mimics an autoinhibitory element of N-WASP into the host cell to activate actin-nucleating factors WASP. The KDEL motif of the bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
encoded cholera toxin mediates cell entry of the cholera toxin.

Potential as leads for drug design
Linear motif mediated protein-protein interactions have shown promise in recent years as novel drug targets. Success stories include the MDM2 motif analog Nutlin-3 andintegrin
Integrins are transmembrane receptors that help cell–cell and cell–extracellular matrix (ECM) adhesion. Upon ligand binding, integrins activate signal transduction pathways that mediate cellular signals such as regulation of the cell cycle, o ...
targeting RGD-mimetic Cilengitide: Nutlin-3 antagonises the interaction of MDM2's SWIB domain with p53
p53, also known as tumor protein p53, cellular tumor antigen p53 (UniProt name), or transformation-related protein 53 (TRP53) is a regulatory transcription factor protein that is often mutated in human cancers. The p53 proteins (originally thou ...
thus stabilising p53 and inducing senescence in cancer cells. Cilengitide inhibits integrin
Integrins are transmembrane receptors that help cell–cell and cell–extracellular matrix (ECM) adhesion. Upon ligand binding, integrins activate signal transduction pathways that mediate cellular signals such as regulation of the cell cycle, o ...
-dependent signaling, causing the disassembly of cytoskeleton
The cytoskeleton is a complex, dynamic network of interlinking protein filaments present in the cytoplasm of all cells, including those of bacteria and archaea. In eukaryotes, it extends from the cell nucleus to the cell membrane and is compos ...
, cellular detachment and the induction of apoptosis
Apoptosis (from ) is a form of programmed cell death that occurs in multicellular organisms and in some eukaryotic, single-celled microorganisms such as yeast. Biochemistry, Biochemical events lead to characteristic cell changes (Morphology (biol ...
in endothelial
The endothelium (: endothelia) is a single layer of squamous endothelial cells that line the interior surface of blood vessels and lymphatic vessels. The endothelium forms an interface between circulating blood or lymph in the lumen and the res ...
and glioma
A glioma is a type of primary tumor that starts in the glial cells of the brain or spinal cord. They are malignant but some are extremely slow to develop. Gliomas comprise about 30% of all brain and central nervous system tumors and 80% of ...
cells. In addition, peptides targeting the Grb2
Growth factor receptor-bound protein 2, also known as Grb2, is an adaptor protein involved in signal transduction/ cell communication. In humans, the GRB2 protein is encoded by the ''GRB2'' gene.
The protein encoded by this gene binds recepto ...
and Crk SH2/ SH3 adaptor domains are also under investigation.
There are at present no drugs on the market specially targeting phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
sites, however, a number of drugs target the kinase
In biochemistry, a kinase () is an enzyme that catalyzes the transfer of phosphate groups from high-energy, phosphate-donating molecules to specific substrates. This process is known as phosphorylation, where the high-energy ATP molecule don ...
domain. This tactic has shown promise in the treatments of various forms of cancer. For example, Stutnet® is a receptor tyrosine kinase
Receptor tyrosine kinases (RTKs) are the high-affinity cell surface receptors for many polypeptide growth factors, cytokines, and hormones. Of the 90 unique tyrosine kinase genes identified in the human genome, 58 encode receptor tyrosine kinas ...
(RTK) inhibitor for treating gastrointestinal cancer, Gleevec
Imatinib, sold under the brand names Gleevec and Glivec (both marketed worldwide by Novartis) among others, is an oral targeted therapy medication used to treat cancer. Imatinib is a small molecule inhibitor targeting multiple tyrosine kinases ...
® specially targets bcr-abl and Sprycel® is a broad-based tyrosine kinase inhibitor whose targets include Bcr-Abl and Src. Cleavage is another process directed by motif recognition with the proteases
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do ...
responsible for cleavage a good drug target. For example, Tritace®, Vasotec
Enalapril, sold under the brand name Vasotec among others, is an ACE inhibitor medication used to treat high blood pressure, diabetic kidney disease, and heart failure. For heart failure, it is generally used with a diuretic, such as furosemi ...
®, Accupril®, and Lotensin® are substrate mimetic Angiotensin
Angiotensin is a peptide hormone that causes vasoconstriction and an increase in blood pressure. It is part of the renin–angiotensin system, which regulates blood pressure. Angiotensin also stimulates the release of aldosterone from the adr ...
converting enzymes inhibitors. Other drugs that target post-translational modifications include Zovirax
Aciclovir, also known as acyclovir, is an antiviral medication. It is primarily used for the treatment of herpes simplex virus infections, chickenpox, and shingles. Other uses include the prevention of cytomegalovirus infections following tran ...
®, an antiviral myristoylation
Myristoylation is a lipidation modification where a myristoyl group, derived from myristic acid, is covalently attached by an amide bond to the alpha-amino group of an ''N''-terminal glycine residue. Myristic acid is a 14-carbon saturated f ...
inhibitor and Farnysyl Transferase inhibitors that block the lipidation modification to a CAAX-box motif.
Recommended further reading:
Computational motif resources
Databases
SLiMs are usually described byregular expression
A regular expression (shortened as regex or regexp), sometimes referred to as rational expression, is a sequence of characters that specifies a match pattern in text. Usually such patterns are used by string-searching algorithms for "find" ...
s in the motif literature with the important residues defined based on a combination of experimental, structural and evolutionary evidence. However, high throughput screening such as phage display has seen a large increase in the available information for many motifs classes allowing them to be described with sequence logos. Several diverse repositories currently curate the available motif data. In terms of scope, the Eukaryotic Linear Motif resource (ELM) and MiniMotif Miner Minimotif Miner is a program and database designed to identify minimotifs in any protein. Minimotifs are short, contiguous peptide sequences that are known to have a function in at least one protein. Minimotifs are also called sequence motifs or sh ...
(MnM) represent the two largest motif databases as they attempt to capture all motifs from the available literature. Several more specific and specialised databases also exist, PepCyber and ScanSite focus on smaller subsets of motifs, phosphopeptide binding and important signaling domains respectively. PDZBase focuses solely on PDZ domain ligands. MEROPS
MEROPS is an online database for peptidases (also known as proteases, proteinases and proteolytic enzymes) and their inhibitors. The classification scheme for peptidases was published by Rawlings & Barrett in 1993, and that for protein inhibito ...
and CutDB curate available proteolytic event data including protease specificity and cleavage sites. There has been a large increase in the number of publications describing motif mediated interactions over past decade and as a result a large amount of the available literature remains to be curated. Recent work has created the tool MiMosa to expedite the annotation process and encourage semantically robust motif descriptions.
Discovery tools
SLiMs are short and degenerate and as a result the proteome is littered with stochastically occurring peptides that resemble functional motifs. The biologically relevant cellular partners can easily distinguish functional motifs, however computational tools have yet to reach a level of sophistication where motif discovery can be accomplished with high success rates. Motif discovery tools can be split into two major categories, discovery of novel instance of known functional motifs class and discovery of functional motifs class, however, they all use a limited and overlapping set of attributes to discriminate true and false positives. The main discrimatory attributes used in motif discovery are: * Accessibility – the motif must be accessible for the binding partner. Intrinsic disorder prediction tools (such as IUPred or GlobPlot), domain databases (such asPfam
Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models. The latest version of Pfam, 37.0, was released in June 2024 and contains 21,979 families. It is cur ...
and SMART
''SMart'' was a British CBBC television programme based on art, which began in 1994 and ended in 2009. The programme was recorded at BBC Television Centre in London. Previously it had been recorded in Studio A at Pebble Mill Studios in Birmingha ...
) and experimentally derived structural data (from sources such as PDB) can be used to check the accessibility of predicted motif instances.
* Conservation – the conservation of a motif correlates strongly with functionality and many experimental motifs are seen as islands of strong constraint in regions of weak conservation. Alignment of homologous proteins can be used to calculate conservation metric for a motif.
* Physicochemical properties – Certain intrinsic properties of residues or stretches of amino acids are strong discriminators of functionality, for example, the propensity of a region of disorder to undergo a disorder to order transition.
* Enrichment in groupings of similar proteins – Motif often evolve convergently to carry out similar tasks in different proteins such as mediating binding to a specific partner or targeting proteins to a particular subcellular localisation. Often in such cases these grouping the motif occurs more often than is expected by chance and can be detected by searching for enriched motifs.
Novel functional motifs instances
The Eukaryotic Linear Motif resource (ELM) andMiniMotif Miner Minimotif Miner is a program and database designed to identify minimotifs in any protein. Minimotifs are short, contiguous peptide sequences that are known to have a function in at least one protein. Minimotifs are also called sequence motifs or sh ...
(MnM) both provide servers to search for novel instance of known functional motifs in protein sequences. SLiMSearch allows similar searches on a proteome-wide scale.
Novel functional motifs class
More recently computational methods have been developed that can identify new Short Linear Motifs de novo. Interactome-based tools rely on identifying a set of proteins that are likely to share a common function, such as binding the same protein or being cleaved by the same peptidase. Two examples of such software are DILIMOT and SLiMFinder. Anchor and α-MoRF-Pred use physicochemical properties to search for motif-like peptides in disordered regions (termed MoRFs, among others). ANCHOR identifies stretches of intrinsically disordered regions that cannot form favorable intrachain interactions to fold without additional stabilising energy contributed by a globular interaction partner. α-MoRF-Pred uses the inherent propensity of many SLiM to undergo a disorder to order transition upon binding to discover α-helical forming stretches within disordered regions. MoRFPred and MoRFchibi SYSTEM are SVM based predictors which utilize multiple features including local sequence physicochemical properties, long stretches of disordered regions and conservation in their predictions. SLiMPred is neural network–based method for the de novo discovery of SLiMs from the protein sequence. Information about the structural context of the motif (predicted secondary structure, structural motifs, solvent accessibility, and disorder) are used during the predictive process. Importantly, no previous knowledge about the protein (i.e., no evolutionary or experimental information) is required.References
External links
Pawsons Lab Resource on motif-binding domains
SLiM databases
Eukaryotic Linear Motif Database
MiniMotif Miner
PepCyber
ScanSite
SLiM discovery tools
ANCHOR
DiLiMot
Eukaryotic Linear Motif Database
MiniMotif Miner
SLiMSuite
: *
SLiMPred
*
*
*
ScanSite
{{MotifBindingDomains Protein structural motifs Protein domains