Membrane Topology on:
[Wikipedia]
[Google]
[Amazon]
Topology of a 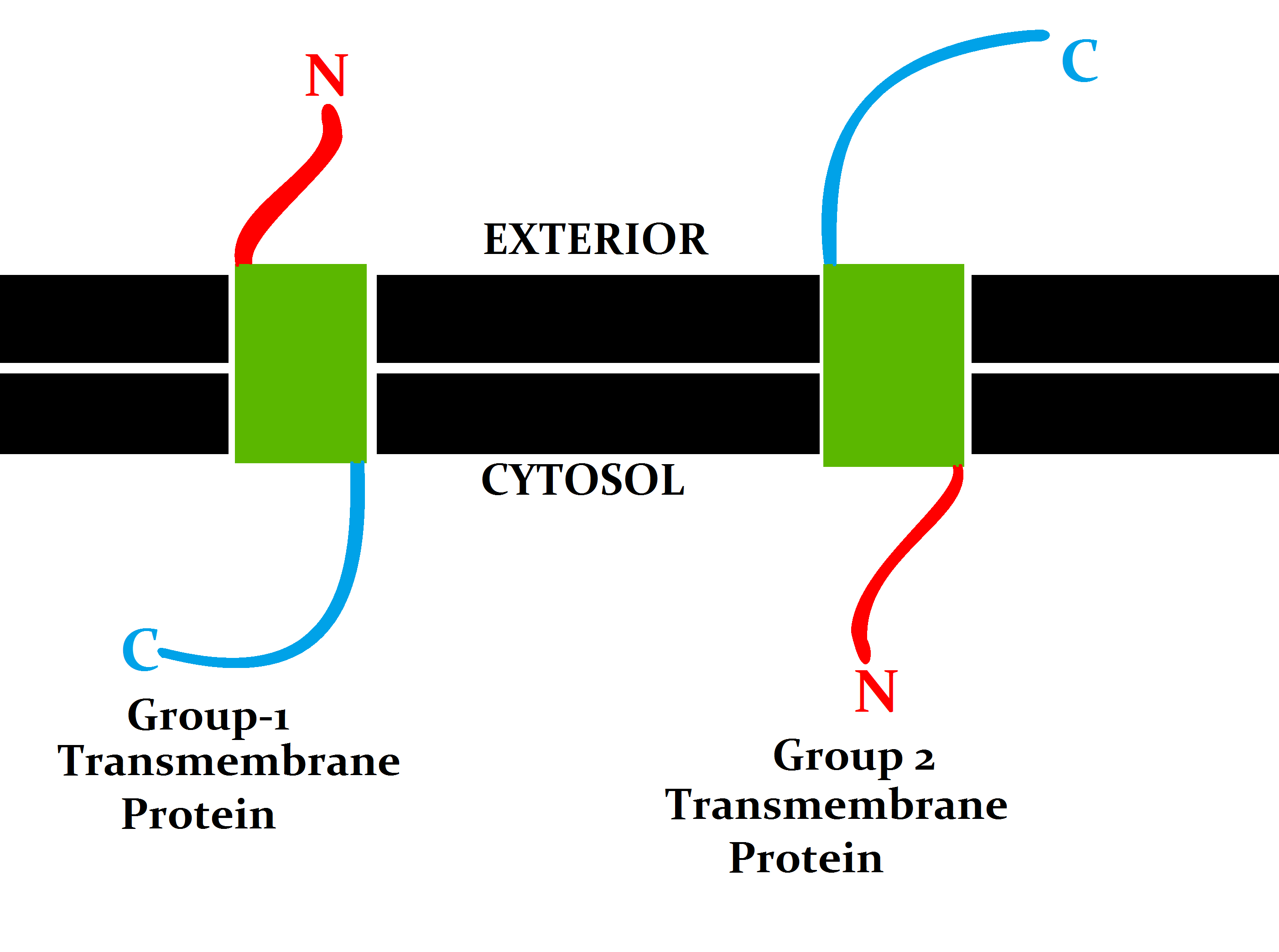 Several databases provide experimentally determined topologies of membrane proteins. They include
Several databases provide experimentally determined topologies of membrane proteins. They include
transmembrane protein
A transmembrane protein is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently un ...
refers to locations of N- and C-termini of membrane-spanning polypeptide chain with respect to the inner or outer sides of the biological membrane
A biological membrane, biomembrane or cell membrane is a selectively permeable membrane that separates the interior of a cell from the external environment or creates intracellular compartments by serving as a boundary between one part of th ...
occupied by the protein.
 Several databases provide experimentally determined topologies of membrane proteins. They include
Several databases provide experimentally determined topologies of membrane proteins. They include Uniprot
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived fro ...
, TOPDB, OPM, and ExTopoDB. There is also a database of domains located conservatively on a certain side of membranes, TOPDOM.
Several computational methods were developed, with a limited success, for predicting transmembrane alpha-helices and their topology. Pioneer methods utilized the fact that membrane-spanning regions contain more hydrophobic residues than other parts of the protein, however applying different hydrophobic scales altered the prediction results. Later, several statistical methods were developed to improve the topography prediction and a special alignment method was introduced. According to the positive-inside rule, cytosolic loops near the lipid bilayer contain more positively-charged amino acids. Applying this rule resulted in the first topology prediction methods. There is also a negative-outside rule in transmembrane alpha-helices from single-pass proteins, although negatively charged residues are rarer than positively charged residues in transmembrane segments of proteins. As more structures were determined, machine learning algorithms appeared. Supervised learning
In machine learning, supervised learning (SL) is a paradigm where a Statistical model, model is trained using input objects (e.g. a vector of predictor variables) and desired output values (also known as a ''supervisory signal''), which are often ...
methods are trained on a set of experimentally determined structures, however, these methods highly depend on the training set. Unsupervised learning
Unsupervised learning is a framework in machine learning where, in contrast to supervised learning, algorithms learn patterns exclusively from unlabeled data. Other frameworks in the spectrum of supervisions include weak- or semi-supervision, wh ...
methods are based on the principle that topology depends on the maximum divergence of the amino acid distributions in different structural parts. It was also shown that locking a segment location based on prior knowledge about the structure improves the prediction accuracy. This feature has been added to some of the existing prediction methods. The most recent methods use consensus prediction (i.e. they use several algorithms to determine the final topology) and automatically incorporate previously determined experimental informations. HTP database provides a collection of topologies that are computationally predicted for human transmembrane proteins.
Discrimination of signal peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16–30 amino acids long) present at the ...
s and transmembrane segments is an additional problem in topology prediction treated with a limited success by different methods. Both signal peptides and transmembrane segments contain hydrophobic regions which form α-helices. This causes the cross-prediction between them, which is a weakness of many transmembrane topology predictors. By predicting signal peptides and transmembrane helices simultaneously (Phobius), the errors caused by cross-prediction are reduced and the performance is substantially increased. Another feature used to increase the accuracy of the prediction is the homology (PolyPhobius).”
It is also possible to predict beta-barrel membrane proteins' topology.
See also
*Endomembrane system
The endomembrane system is composed of the different membranes (endomembranes) that are suspended in the cytoplasm within a eukaryotic cell. These membranes divide the cell into functional and structural compartments, or organelles. In eukaryote ...
* Integral membrane protein
An integral, or intrinsic, membrane protein (IMP) is a type of membrane protein that is permanently attached to the biological membrane. All transmembrane proteins can be classified as IMPs, but not all IMPs are transmembrane proteins. IMPs comp ...
* Protein topology
* Structural biology
Structural biology deals with structural analysis of living material (formed, composed of, and/or maintained and refined by living cells) at every level of organization.
Early structural biologists throughout the 19th and early 20th centuries we ...
* Transmembrane domain
A transmembrane domain (TMD, TM domain) is a membrane-spanning protein domain. TMDs may consist of one or several alpha-helices or a transmembrane beta barrel. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in ...
References
{{DEFAULTSORT:Membrane Topology Integral membrane proteins Membrane biology Molecular topology