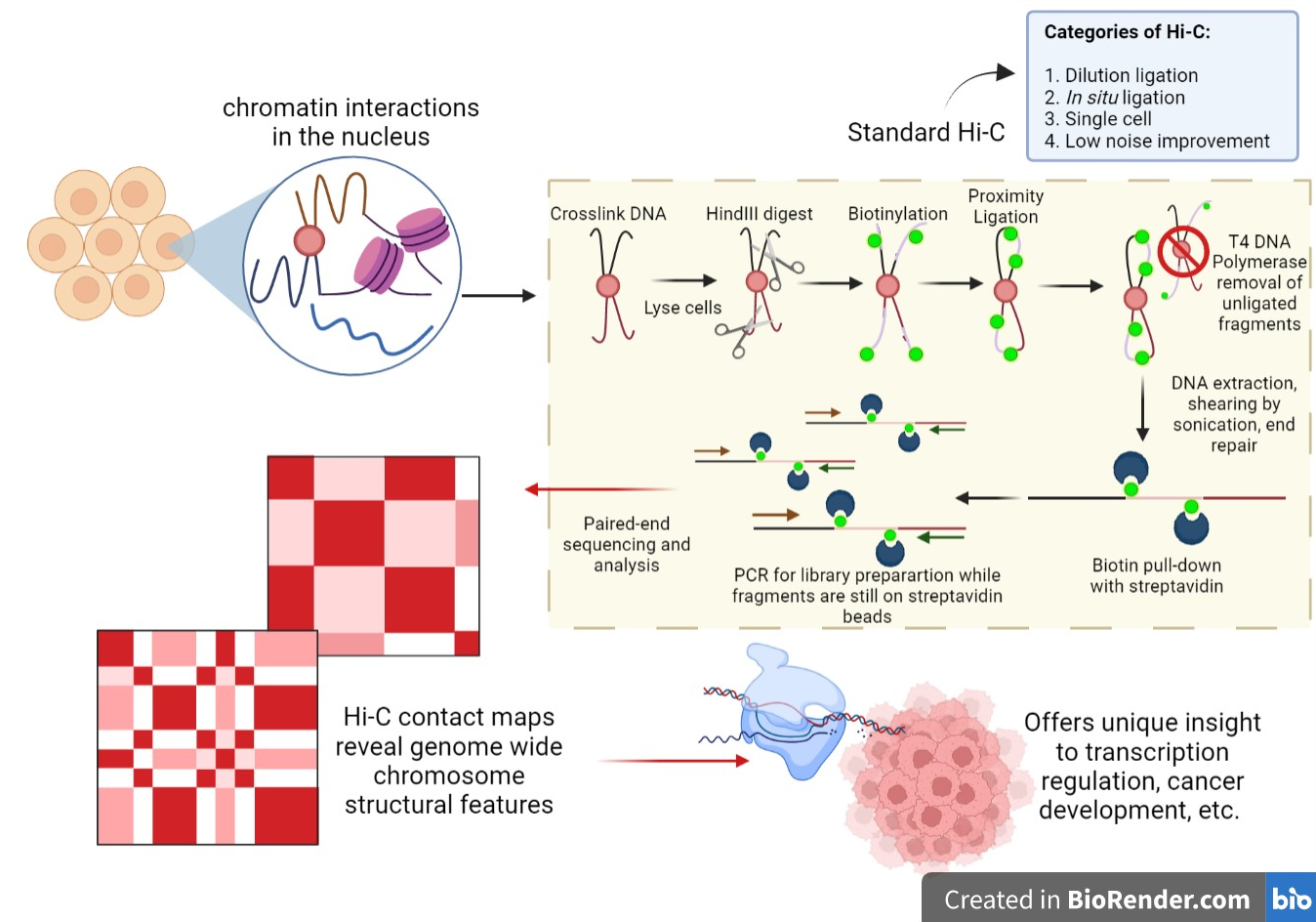
]
Hi-C (or standard Hi-C) is a high-throughput
Genomics, genomic and
epigenomic technique first described in 2009 by Lieberman-Aiden et al. to
capture chromatin conformation.
In general, Hi-C is considered as a derivative of a series of
chromosome conformation capture
Chromosome conformation capture techniques (often abbreviated to 3C technologies or 3C-based methods) are a set of molecular biology methods used to analyze the spatial organization of chromatin in a cell. These methods quantify the number of int ...
technologies, including but not limited to 3C (chromosome conformation capture), 4C (chromosome conformation capture-on-chip/circular chromosome conformation capture), and 5C (chromosome conformation capture carbon copy).
Hi-C comprehensively detects genome-wide chromatin interactions in the
cell nucleus by combining 3C and
next-generation sequencing (NGS) approaches and has been considered as a qualitative leap in C-technology (chromosome conformation capture-based technologies) development and the beginning of 3D genomics.
Similar to the classic 3C technique, Hi-C measures the frequency (as an average over a cell population) at which two DNA fragments physically associate in 3D space, linking chromosomal structure directly to the genomic sequence.
The general procedure of Hi-C involves first crosslinking chromatin material using
formaldehyde
Formaldehyde ( , ) ( systematic name methanal) is a naturally occurring organic compound with the formula and structure . The pure compound is a pungent, colourless gas that polymerises spontaneously into paraformaldehyde (refer to section ...
.
Then, the chromatin is solubilized and fragmented, and interacting
loci
Locus (plural loci) is Latin for "place". It may refer to:
Entertainment
* Locus (comics), a Marvel Comics mutant villainess, a member of the Mutant Liberation Front
* ''Locus'' (magazine), science fiction and fantasy magazine
** '' Locus Award ...
are
re-ligated together to create a genomic library of
chimeric DNA molecules.
The relative abundance of these chimeras, or ligation products, is correlated to the probability that the respective chromatin fragments interact in 3D space across the cell population.
While 3C focuses on the analysis of a set of predetermined genomic loci to offer “one-versus-some” investigations of the conformation of the chromosome regions of interest, Hi-C enables “all-versus-all” interaction profiling by labeling all fragmented chromatin with a biotinylated
nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecul ...
before ligation.
As a result,
biotin-marked ligation junctions can be purified more efficiently by
streptavidin
Streptavidin is a 66.0 (tetramer) kDa protein purified from the bacterium ''Streptomyces avidinii''. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin (also known as vitamin B7 or vitamin H). With a dissociation c ...
-coated magnetic beads, and chromatin interaction data can be obtained by direct sequencing of the Hi-C library.
Analyses of Hi-C data not only reveal the overall genomic structure of mammalian
chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins ar ...
s, but also offer insights into the biophysical properties of chromatin as well as more specific, long-range contacts between distant genomic elements (e.g. between
gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
s and
regulatory elements
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and vir ...
).
In recent years, Hi-C has found its application in a wide variety of biological fields, including
cell growth and
division
Division or divider may refer to:
Mathematics
*Division (mathematics), the inverse of multiplication
*Division algorithm, a method for computing the result of mathematical division
Military
* Division (military), a formation typically consisting ...
,
transcription regulation
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA ( transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from al ...
,
fate determination, development, disease, and
genome evolution
Genome evolution is the process by which a genome changes in structure (sequence) or size over time. The study of genome evolution involves multiple fields such as structural analysis of the genome, the study of genomic parasites, gene and ancient ...
.
By combining Hi-C data with other datasets such as genome-wide maps of chromatin modifications and gene expression profiles, the functional roles of chromatin conformation in genome regulation and stability can also be delineated.
History
At its inception, Hi-C was a low-resolution, high-noise technology that was only capable of describing chromatin interaction regions within a bin size of 1 million
base pairs (Mb).
The Hi-C library also required several days to construct,
and the datasets themselves were low in both output and reproducibility.
Nevertheless, Hi-C data offered new insights for chromatin conformation as well as nuclear and genomic architectures, and these prospects motivated scientists to put efforts to modify the technique over the past decade.
Between 2012 and 2015, several modifications to the Hi-C protocol have taken place, with 4-cutter digestion
or adapted deeper sequencing depth to obtain higher resolution.
The use of
restriction endonucleases
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class ...
that cut more frequently, or
DNaseI
Deoxyribonuclease I (usually called DNase I), is an endonuclease of the DNase family coded by the human gene DNASE1.
DNase I is a nuclease that cleaves DNA preferentially at phosphodiester linkages adjacent to a pyrimidine nucleotide, yielding ...
and
Micrococcal nuclease
Micrococcal nuclease (, ''S7 Nuclease'', ''MNase'', ''spleen endonuclease'', ''thermonuclease'', ''nuclease T'', ''micrococcal endonuclease'', ''nuclease T, ''staphylococcal nuclease'', ''spleen phosphodiesterase'', ''Staphylococcus aureus nucle ...
s also significantly increased the resolution of the method.
More recently (2017), Belaghzal et al. described a Hi-C 2.0 protocol that was able to achieve kilobase (kb) resolution.
The key adaptation to the base protocol was the removal of the
SDS solubilization step after digestion to preserve nuclear structure and prevent random ligation between fragmented chromatin by ligation within the intact nuclei, which formed the basis of in situ Hi-C.
In 2021, Hi-C 3.0 was described by Lafontaine et al., with higher resolution achieved by enhancing crosslinking with formaldehyde followed by disuccinimidyl glutarate (DSG).
While formaldehyde captures the
amino
In chemistry, amines (, ) are compounds and functional groups that contain a basic nitrogen atom with a lone pair. Amines are formally derivatives of ammonia (), wherein one or more hydrogen atoms have been replaced by a substituent su ...
and
imino
In organic chemistry, an imine ( or ) is a functional group or organic compound containing a carbon–nitrogen double bond (). The nitrogen atom can be attached to a hydrogen or an organic group (R). The carbon atom has two additional single bon ...
groups of both proteins and DNA, the NHS-esters in DSG react with primary amines on proteins and can capture amine-amine interactions.
These updates to the base protocol allowed the scientists to look at more detailed conformational structures such as chromosomal compartment and
topologically associating domain
A topologically associating domain (TAD) is a self-interacting genomic region, meaning that DNA sequences within a TAD physically interact with each other more frequently than with sequences outside the TAD. The median size of a TAD in mouse cells ...
s (TADs), as well as high-resolution conformational features such as DNA loops.
To date, a variety of derivatives of Hi-C have already emerged, including in situ Hi-C, low Hi-C, SAFE Hi-C, and Micro-C, with distinctive features related to different aspects of standard Hi-C, but the basic principle has remained the same.
Traditional Hi-C
First published by Lieberman-Aiden, et al. in 2009, the outline of the classical Hi-C workflow is as follows: cells are cross-linked with formaldehyde; chromatin is digested with a restriction enzyme that generates a
5’ overhang; the 5’ overhang is filled with biotinylated bases and the resulting blunt-ended DNA is ligated.
The ligation products, with biotin at the junction, are selected for using streptavidin and further processed to prepare a library ready for subsequent sequencing efforts.
The pairwise interactions that Hi-C can capture across the genome are immense and so it is important to analyze an appropriately large sample size, in order to capture unique interactions that may only be observed in a minority of the general population.
To obtain a high complexity library of ligation products that will ensure high resolution and depth of data, a sample of 20–25 million cells is required as input for Hi-C.
Primary human samples, which may be available only in fewer cell numbers, could be used for standard Hi-C library preparation with as low as 1–5 million cells.
However, using such a low input of cells may be associated with low library complexity which results in a high percentage of duplicate reads during library preparation.
Standard Hi-C gives data on pairwise interactions at the resolution of 1 to 10 Mb, requires high sequencing depth and the protocol takes around 7 days to complete.
Formaldehyde cross-linking
 Cell and nuclear membranes
Cell and nuclear membranes are highly permeable to formaldehyde.
Formaldehyde cross-linking is frequently employed for the detection and quantification of DNA-protein and protein-protein interactions.
Of interest in the context of Hi-C, and all 3C-based methods, is the ability of formaldehyde to capture cis chromosomal interactions between distal segments of chromatin.
It does so by forming
covalent links between spatially adjacent chromatin segments. Formaldehyde can react with
macromolecule
A macromolecule is a very large molecule important to biophysical processes, such as a protein or nucleic acid. It is composed of thousands of covalently bonded atoms. Many macromolecules are polymers of smaller molecules called monomers. ...
s in two steps: first it reacts with a
nucleophilic
In chemistry, a nucleophile is a chemical species that forms bonds by donating an electron pair. All molecules and ions with a free pair of electrons or at least one pi bond can act as nucleophiles. Because nucleophiles donate electrons, they a ...
group on a DNA base for example, and forms a methylol adduct, which is then converted to a
Schiff base
In organic chemistry, a Schiff base (named after Hugo Schiff) is a compound with the general structure ( = alkyl or aryl, but not hydrogen). They can be considered a sub-class of imines, being either secondary ketimines or secondary aldimine ...
.
In the second step, the Schiff base, which can decompose rapidly, forms a
methylene bridge
In organic chemistry, a methylene bridge, methylene spacer, or methanediyl group is any part of a molecule with formula ; namely, a carbon atom bound to two hydrogen atoms and connected by single bonds to two other distinct atoms in the rest of ...
with another functional group on another molecule.
It can also make this methylene bridge with a small molecule in solution such as
glycine
Glycine (symbol Gly or G; ) is an amino acid that has a single hydrogen atom as its side chain. It is the simplest stable amino acid ( carbamic acid is unstable), with the chemical formula NH2‐ CH2‐ COOH. Glycine is one of the proteinog ...
, which is used in excess to quench formaldehyde in Hi-C.
Quenchers can typically exert an effect on formaldehyde from outside the cell.
A key feature of this two-step formaldehyde crosslinking reaction is that all the reactions are reversible, which is vital for chromatin capture.
Crosslinking is a pivotal step of the chromatin capture workflow as the functional readout of the technique is the frequency at which two genomic regions are crosslinked to each other.
Thus, the standardization of this step is important and for that, one must consider potential sources of variation.
Presence of serum, which contains a high concentration of protein, in culture media can decrease the effective concentration of formaldehyde available for chromatin crosslinking, by sequestering it in the culture media.
Therefore in cases where serum is used in culture, it should be removed for the crosslinking step.
The nature of cells, i.e., whether they are suspension or adherent, is also a pertinent consideration for the crosslinking step.
Adherent cells bind to surfaces with the help of molecular mechanisms of
cytoskeleton
The cytoskeleton is a complex, dynamic network of interlinking protein filaments present in the cytoplasm of all cells, including those of bacteria and archaea. In eukaryotes, it extends from the cell nucleus to the cell membrane and is comp ...
s.
It has been shown that there is a link between cytoskeleton-maintained nucelear and cellular morphology which, if altered, may negatively impact global nuclear organization.
Adherent cells therefore, should be crosslinked while still attached to their culture surface.
Lysis, Restriction Digest and Biotinylation
Cells are lysed on ice with cold
hypotonic
In chemical biology, tonicity is a measure of the effective osmotic pressure gradient; the water potential of two solutions separated by a partially-permeable cell membrane. Tonicity depends on the relative concentration of selective membrane-i ...
buffer containing
sodium chloride
Sodium chloride , commonly known as salt (although sea salt also contains other chemical salts), is an ionic compound with the chemical formula NaCl, representing a 1:1 ratio of sodium and chloride ions. With molar masses of 22.99 and 35 ...
,
Tris-HCl
Tris, or tris(hydroxymethyl)aminomethane, or known during medical use as tromethamine or THAM, is an organic compound with the formula (HOCH2)3CNH2, one of the twenty Good's buffers. It is extensively used in biochemistry and molecular biology as ...
at pH 8.0, and
non-ionic
An ion () is an atom or molecule with a net electrical charge.
The charge of an electron is considered to be negative by convention and this charge is equal and opposite to the charge of a proton, which is considered to be positive by conven ...
detergent
A detergent is a surfactant or a mixture of surfactants with cleansing properties when in dilute solutions. There are a large variety of detergents, a common family being the alkylbenzene sulfonates, which are soap-like compounds that are m ...
IGEPAL CA-630
IGEPAL CA-630 is a nonionic, non-denaturing detergent. Its official IUPAC name is octylphenoxypolyethoxyethanol. IGEPAL is a registered trademark of Rhodia.
IGEPAL CA-630 is sold by Sigma-Aldrich and is claimed to be a "chemically indistinguisha ...
, supplemented with
protease inhibitors
Protease inhibitors (PIs) are medications that act by interfering with enzymes that cleave proteins. Some of the most well known are antiviral drugs widely used to treat HIV/AIDS and hepatitis C. These protease inhibitors prevent viral repli ...
.
The protease inhibitors and incubation on ice help preserve the integrity of crosslinked chromatin complexes from endogenous proteases.
The lysis step helps to release the nucleic material from the cells.
Following cell lysis, chromatin is solubilized with dilute SDS in order to remove proteins that have not been crosslinked and to open chromatin and make it more accessible for subsequent restriction endonuclease-mediated digestion.
If the incubation with SDS exceeds the recommended 10 minutes, the formaldehyde crosslinks can be reversed and so the incubation with SDS must be immediately followed by an incubation on ice.
A non-ionic detergent called
Triton X-100
Triton X-100 (''n'') is a nonionic surfactant that has a hydrophilic polyethylene oxide chain (on average it has 9.5 ethylene oxide units) and an aromatic hydrocarbon lipophilic or hydrophobic group. The hydrocarbon group is a 4-( 1,1,3,3-tetra ...
is used to quench SDS in order to prevent
enzyme denaturation
In biochemistry, denaturation is a process in which proteins or nucleic acids lose the quaternary structure, tertiary structure, and secondary structure which is present in their native state, by application of some external stress or compound ...
in the next step.
Any restriction enzyme that generates a 5’ overhang, such as
HindIII
''Hin''dIII (pronounced "Hin D Three") is a type II site-specific deoxyribonuclease restriction enzyme isolated from '' Haemophilus influenzae'' that cleaves the DNA palindromic sequence AAGCTT in the presence of the cofactor Mg2+ via hydroly ...
can be used to digest the now accessible chromatin overnight.
This 5’ overhang provides the template required by the
Klenow fragment
The Klenow fragment is a large protein fragment produced when DNA polymerase I from ''E. coli'' is enzymatically cleaved by the protease subtilisin. First reported in 1970, it retains the 5' → 3' polymerase activity and the 3’ → 5’ exonuc ...
of
DNA Polymerase I
DNA polymerase I (or Pol I) is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase (and the first known of any kind of polymerase). It was initial ...
to add biotinylated CTP or ATP to the digested ends of chromatin.
This step allows for the selection of Hi-C ligation products for library preparation.
Proximity Ligation
A dilution ligation is performed on DNA fragments that are still crosslinked to one another in order to favor the intramolecular ligation of fragments within the same chromatin complex instead of ligation events between fragments across different complexes.
Since this ligation step occurs between blunt-ended DNA fragments (since the sticky ends have been filled in with biotin-labeled bases), the reaction is allowed to go on for up to 4 hours to make up for its inherent inefficiency.
As a result of proximity ligation, the terminal HindIII sites are lost and an NheI site is generated.
Biotin Removal, DNA Shearing, Size Selection and End Repair
The biotin-labeled ligation products can be purified using
phenol-chloroform DNA extraction.
To remove any fragments with biotin-labeled ends that have not been ligated, T4 DNA Polymerase with 3’ to 5’
exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is t ...
activity is used to remove nucleotides from the ends of such fragments.
This step ensures that none of these unligated fragments are selected for library preparation.
The reaction is stopped with
EDTA
Ethylenediaminetetraacetic acid (EDTA) is an aminopolycarboxylic acid with the formula H2N(CH2CO2H)2sub>2. This white, water-soluble solid is widely used to bind to iron (Fe2+/Fe3+) and calcium ions (Ca2+), forming water-soluble complexes ev ...
and the DNA is purified once again using phenol-chloroform DNA extraction.
The ideal size of DNA fragments for the sequencing library depends on the sequencing platform that will be used.
DNA can first be sheared to fragments around 300–500 bp long using
sonication
A sonicator at the Weizmann Institute of Science during sonicationSonication is the act of applying sound energy to agitate particles in a sample, for various purposes such as the extraction of multiple compounds from plants, microalgae and seawe ...
.
Fragments of this size are suitable for high-throughput sequencing.
Following sonication, fragments can be size selected using AMPure XP beads from
Beckman Coulter
Beckman Coulter Inc. is a Danaher Corporation company that develops, manufactures, and markets products that simplify, automate and innovate complex biomedical testing. It operates in two industries: Diagnostics and Life Sciences. For more than ...
to obtain ligation products with a size distribution between 150–300 bp.
This is the optimal fragment size window for HiSeq cluster formation.
DNA shearing causes asymmetric DNA breaks and must be repaired before biotin pulldown and sequencing adaptor ligation.
This is achieved by using a combination of enzymes that fill in 5’ overhangs, and add 5’ phosphate groups and adenylate to the 3’ ends of fragments to allow for ligation of sequencing adaptors.
Biotin Pull-Down
Using an excess of streptavdin beads, such as the My-One C1 streptavidin bead solution from
Dynabeads Dynabeads are superparamagnetic spherical polymer particles with a uniform size and a consistent, defined surface for the adsorption or coupling of various bioreactive molecules or cells.
Description
Dynabeads were developed after John Ugelstad m ...
, biotinylated Hi-C ligation products can be pulled-down and enriched for.
Ligation of the Illumina paired-end adapters is performed while the DNA fragments are bound to the streptavidin beads.
Adsorption to the beads increases efficiency of the ligation of these blunt-ended DNA fragments to the adaptors, as it decreases their mobility.
Library Preparation and Sequencing
After the ligation of the adaptors is complete,
PCR PCR or pcr may refer to:
Science
* Phosphocreatine, a phosphorylated creatine molecule
* Principal component regression, a statistical technique
Medicine
* Polymerase chain reaction
** COVID-19 testing, often performed using the polymerase chain r ...
amplification of the library is performed.
The PCR step can introduce high number of duplicates in a low complexity Hi-C ligation product sample as a result of over-amplification.
This results in very few interactions being captured and often times, this is because the input sample size had a low amount of cells.
It is important to titrate the number of cycles required to get at least 50 ng of Hi-C library DNA for sequencing.
Fewer the cycle number, the better so that there are no PCR artifacts (such as off-target amplicons, non-specificity, etc.).
The ideal range of PCR cycles is 9–15 and it is more ideal to pool multiple PCR reactions to get enough DNA for sequencing, than to increase the number of cycles for one PCR reaction.
The PCR products are purified again using AMPure beads to remove
primer dimer A primer dimer (PD) is a potential by-product in the polymerase chain reaction (PCR), a common biotechnological method. As its name implies, a PD consists of two primer molecules that have attached ( hybridized) to each other because of strings of ...
s and then quantified before being sequenced.
Regions of chromatin that interact with each other are then identified by paired-end sequencing of the biotinylated, ligated products.
Any platform that can allow for the ligated fragments to be sequenced across the NheI junction (
Roche
F. Hoffmann-La Roche AG, commonly known as Roche, is a Swiss multinational healthcare company that operates worldwide under two divisions: Pharmaceuticals and Diagnostics. Its holding company, Roche Holding AG, has shares listed on the SIX S ...
454) or by paired-end or mate-paired reads (
Illumina GA and
HiSeq
Illumina, Inc. is an American biotechnology company, headquartered in San Diego, California. Incorporated on April 1, 1998, Illumina develops, manufactures, and markets integrated systems for the analysis of genetic variation and biological fun ...
platforms) would be suitable for Hi-C.
Before high-throughput sequencing, the quality of the library should be verified using
Sanger sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Frede ...
, wherein the long sequencing read will read through the biotin junction.
Thirty-six or 50 bp reads are sufficient to identify most chromatin interacting pairs using Illumina paired-end sequencing.
Since the average size of fragments in the library is 250 bp, 50bp paired-end reads have been found to be optimum for Hi-C library sequencing.
Quality Control of Hi-C Libraries
There are several pressure points throughout the workflow of Hi-C sample preparation that are well documented and reported.
DNA at various stages can be run on 0.8% agarose gels to assay the size distribution of fragments.
This is particularly important after shearing of size selection steps.
Degradation of DNA can also be monitored as smears appearing as a result under low molecular weight products on gels.
Degradation can occur due to not adding sufficient protease inhibitors during lysis, endogenous nuclease activity or thermal degradation due to incorrect icing.
3C PCR reactions can be performed to test for the formation of proximity ligation products.
Variants
Standard Hi-C has a high input cell number cost, requires deep sequencing, generates low-resolution data, and suffers from formation of redundant molecules that contribute to low complexity libraries when cell numbers are low.
To combat these issues in order to be able to apply this technique in contexts where cell number is a limiting factor, for example, with primary human cell work, several Hi-C variants have been developed since the first conceptualization of Hi-C.
The four main classes under which Hi-C variants fall under are: dilution ligation, in situ ligation, single cell, and low noise improvement systems.
Standard Hi-C is a type of dilution ligation and other dilution ligation include DNase Hi-C and Capture Hi-C.
In contrast to standard and Capture Hi-C, DNase Hi-C requires only 2–5 million cells as input, uses DNaseI for chromatin fragmentation and employs an in-gel dilution proximity ligation.
The use of DNaseI has been shown to greatly improve efficiency and resolution of Hi-C.
Capture Hi-C is a genome-wide assaying technique to look at chromatin interactions of specific loci using a
hybridization
Hybridization (or hybridisation) may refer to:
*Hybridization (biology), the process of combining different varieties of organisms to create a hybrid
*Orbital hybridization, in chemistry, the mixing of atomic orbitals into new hybrid orbitals
*Nu ...
-based capture of targeted genomic regions.
It was first developed by Mifsud et al. to map long-range
promoter
Promoter or Promotor may refer to:
Art, entertainment, and media
* ''The Promoter'' (1952), also known as ''The Card''
* ''The Promoter'' (2012 film)
Professions
* Promoter (entertainment), one who makes arrangements for events or markets them ...
contacts in human cells by generating a biotinylated RNA bait library that targeted 21,841 promoter regions.
These variants, in addition to others (described below), represent modifications to the foundational technique of standard Hi-C and address and alleviate one or more limitations of the original method.
''In situ'' Hi-C
First described by Rao et al., in situ Hi-C combines standard Hi-C with nuclear ligation assay, i.e., proximity ligation performed in intact nuclei.
The protocol is similar to standard Hi-C in terms of the basic workflow outline but differs in other ways.
In situ Hi-C requires 2 to 5 million cells compared to the ideal 20 to 25 million required for standard Hi-C and it requires only 3 days to complete the protocol versus 7 days for standard Hi-C.
Furthermore, proximity ligation does not take place in solution like in standard Hi-C, decreasing the frequency of random, biologically-irrelevant contacts and ligations, as indicated by the lower frequency of mitochondrial and nuclear DNA contacts in captured biotinylated DNA.
This is achieved by leaving the nuclei intact for the ligation step.
Cells are still lysed with a buffer containing Tris-HCl at pH 8.0, sodium chloride, and the detergent IGEPAL CA630 before ligation, but instead of homogenization of the cell lysate, cell nuclei are pelleted after initial lysis to degrade the cell membrane.
After proximity ligation is complete, cell nuclei are incubated for at least 1.5 hours at 68 degrees Celsius to permeabilize the nuclear membrane and release its nuclear contents.
The resolution that can be achieved with in situ Hi-C can be up to 950 to 1000 bp compared to the 1 to 10 Mb resolution of standard Hi-C and the 100 kb resolution of DNase Hi-C.
While standard Hi-C makes use of a 6-bp cutter such as HindIII for the restriction digest step, in situ Hi-C uses a 4-bp cutter such as MboI or its
isoschizomer
Isoschizomers are pairs of restriction enzymes specific to the same recognition sequence. For example, SphI (CGTAC/G) and BbuI (CGTAC/G) are isoschizomers of each other. The first enzyme discovered which recognizes a given sequence is known as th ...
DpnII (which is not sensitive to
CpG methylation) to increase efficiency and resolution (as the restriction sites of MboI and DpnII are more frequently occurring in the genome).
Data between replicates for in situ Hi-C is consistent and highly reproducible, with very less background noise and demonstrating clear chromatin interactions.
It is however possible that some of the captured interactions may not be accurate intermolecular interactions since the nucleus is densely packed with protein and DNA so performing proximity ligations in intact nuclei may pull down confounding interactions that may only form due to the nature of nuclear packaging and not so much unique chromosomal interactions with cellular functional impact.
It also requires an extremely high sequencing depth of around 5 billion paired-end reads per sample to achieve the resolution of data described by Rao et al.
Several techniques that have adapted the concept of in situ Hi-C exist, including Sis Hi-C, OCEAN-C and in situ capture Hi-C.
Described below are two of the most prominent in situ Hi-C based techniques.
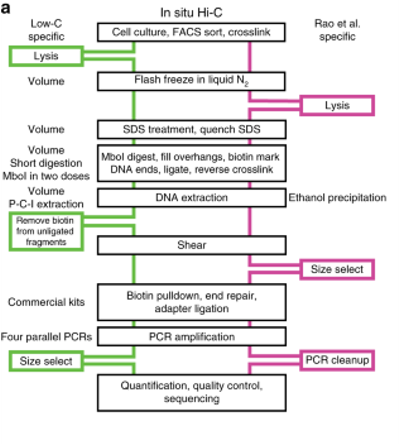
1. Low-C
Low-C is an in situ Hi-C protocol adapted for use on low cell numbers, which is particularly useful in contexts where cell number is a limiting agent, for example, in primary human cell culture.
This method makes use of minor changes, including volumes and concentrations used and the timing and order of certain experimental steps to allow for the generation of high-quality Hi-C libraries from cell numbers as low as 1000 cells.
Despite the potential of generating usable and high resolution data with as few as 1000 cells, Diaz et al. still recommend using at least 1 to 2 million cells if feasible, or if not a minimum of 500 K cells.
Library quality was first assessed on the Illumina MiSeq (2x84 np paired-end reads) platform and once passed quality control criteria (including low PCR duplicates), the library was sequenced on Illumina NextSeq (2x80 bp paired-end).
Overall, this technique circumvents the issue of requiring a high cell number input for Hi-C and the high sequencing depth required to obtain high resolution data, but can only achieve resolutions of up to 5 kb and may not always be reproducible due the variable nature of sample sizes used and the data generated from it.

2. SAFE Hi-C
SAFE Hi-C, or simplified, fast, and economically efficient Hi-C, generates sufficient ligated fragments without amplification for high-throughput sequencing.
In situ Hi-C data that has been published indicates that amplification (at the PCR step for library preparation) introduces distance-dependent amplification bias, which results in a higher noise to signal ratio against genomic distance.
SAFE Hi-C was successfully used to generate an amplification-free, in situ Hi-C ligation library from as low as 250 thousand
K562 cells K562 cells were the first human immortalised myelogenous leukemia cell line to be established. K562 cells are of the Acute erythroid leukemia , erythroleukemia type, and the cell line is derived from a 53-year-old female chronic myelogenous leukemi ...
.
Ligation fragments are anywhere between 200 to 500 bp long, with an average at about 370 bp.
All ligation product libraries were sequenced using the Illumina HiSeq platform (2x150 bp paired-end reads).
Although SAFE Hi-C can be used for a cell input as low as 250 thousand, Niu et al. recommend using 1 to 2 million cells.
Samples produce enough ligates to be sequenced on one-fourth of a lane.
SAFE Hi-C has been demonstrated to increase library complexity due to the removal of PCR duplicates which lower the overall percentage of unique paired reads.
Overall, SAFE Hi-C preserves the integrity of chromosomal interactions while also reducing the need to have high sequencing depth and saving overall cost and labor.
Micro-C
Micro-C is a version of Hi-C that includes a micrococcal nuclease (MNase) digestion step to look at interactions between pairs of
nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamen ...
s, thus enabling resolution of sub-genomic TAD structures at the 1 to 100 nucleosome scale.
It was first developed for use in yeast and was shown to conserve the structural data obtained from a standard Hi-C but with greater signal-to-noise ratio.
When used with
human embryonic stem cells
Embryonic stem cells (ESCs) are pluripotent stem cells derived from the inner cell mass of a blastocyst, an early-stage pre- implantation embryo. Human embryos reach the blastocyst stage 4–5 days post fertilization, at which time they consist ...
and
fibroblast
A fibroblast is a type of biological cell that synthesizes the extracellular matrix and collagen, produces the structural framework ( stroma) for animal tissues, and plays a critical role in wound healing. Fibroblasts are the most common cells of ...
s, 2.6 to 4.5 billion uniquely mapped reads were obtained per sample.
Hsieh et al. analyzed 2.64 billion reads from mouse embryonic stem cells and demonstrated that there was increased power for detecting short-range interactions.

Single cell Hi-C
Hi-C has also been adapted for use with single cells but these techniques require high levels of expertise to perform and are plagued with issues such as low data quality, coverage, and resolution.
Data Analysis
The chimeric DNA ligation products generated by Hi-C represent pairwise chromatin interactions or physical 3D contacts within the nucleus,
and can be analyzed by a variety of downstream approaches. Briefly, deep sequencing data is used to build unbiased genome-wide chromatin interaction maps.
Then several different methods can be employed to analyze these maps to identify chromosomal structural patterns and their biological interpretations. Many of these data analysis approaches also apply to 3C-sequencing or other equivalent data.
Read Mapping
Hi-C data produced by deep sequencing is in the form of a traditional
FASTQ file, and the reads can be aligned to the genome of interest using
sequence alignment software
This list of sequence alignment software is a compilation of software tools and web portals used in pairwise sequence alignment and multiple sequence alignment. See structural alignment software for structural alignment of proteins.
Database searc ...
(e.g.
Bowtie
The bow tie is a type of necktie. A modern bow tie is tied using a common shoelace knot, which is also called the bow knot for that reason. It consists of a ribbon of fabric tied around the collar of a shirt in a symmetrical manner so that ...
,
bwa,
etc.).
Because Hi-C ligation products may span hundreds of megabases and may bridge loci on different chromosomes,
Hi-C read alignment is often chimeric in the sense that different parts of a read may be aligned to loci distant apart, possibly in different orientations. Long-read aligners (e.g. minimap2) often support chimeric alignment and can be directly applied to long-read Hi-C data. Short-read Hi-C alignment is more challenging.
Notably, Hi-C generates ligation junctions of varying sizes, but the exact position of the ligation site is not measured.
To circumvent this problem, iterative mapping
is used to avoid the search for the junction site before being able to split the reads into two and mapping them separately to identify the interaction pairs. The idea behind iterative mapping is to map as short a sequence as possible to ensure unique identification of interaction pairs before reaching the junction site.
As a result, 25-bp long reads starting from the 5’ end are mapped to the genome at first, and reads that do not uniquely map to a single loci are extended by an additional 5 bp and then re-mapped.
This process is repeated till all reads uniquely map, or till the reads are extended to their entirety.
Only paired end reads with each side uniquely mapped to a single genomic loci are kept.
All other paired end reads are discarded.
Several variations of read mapping techniques are implemented in many bioinformatics pipelines, such as ICE,
HiC-Pro,
HIPPIE,
HiCUP,
and TADbit,
to map two portions of a paired end read separately, in the case that the two portions match distinct genomic positions, thus addressing the challenge where reads span the ligation junctions.
With increased read length, more recent pipelines (e.g. Juicer
and the 4D-Nucleosome Data Portal) often align short Hi-C reads with an alignment algorithm capable of chimeric alignment, such as bwa-mem, chromap
an
dragmap This procedure calls alignment once and is simpler than iterative mapping.
Fragment assignment and filtering
The mapped reads are then each assigned a single genomic alignment location according to its 5’ mapped position in the genome.
For each read pair, a location is assigned to only one of the
restriction fragment
A restriction fragment is a DNA fragment resulting from the cutting of a DNA strand by a restriction enzyme (restriction endonucleases), a process called restriction. Each restriction enzyme is highly specific, recognising a particular short DNA s ...
s, thus should fall in close proximity to a
restriction site Restriction sites, or restriction recognition sites, are located on a DNA molecule containing specific (4-8 base pairs in length) sequences of nucleotides, which are recognized by restriction enzymes. These are generally palindromic sequences (bec ...
and less than the maximum molecule length away.
Reads mapped more than the maximum molecule length away from the closest restriction sites are the results of physical breakage of the chromatin or non-canonical nuclease activities.
Because these reads also instruct information on chromatin interactions, they are not discarded, but appropriate filtering must take place after assigning genomic locations to remove technical noise in the dataset.
Depending on whether the read pair falls within the same or different restriction fragments, different filtering criteria are applied. If the paired reads map to the same restriction fragment, they likely represent un-ligated dangling ends or circularized fragments that are uninformative, and are therefore removed from the dataset.
These reads could also represent PCR artifacts, undigested chromatin fragments, or simply, reads with low alignment quality.
Whatever their origin, reads mapped to the same fragment are considered “spurious signals”
and are typically discarded before downstream processing.
The remaining paired reads mapped to distinct restriction fragments are also filtered to discard identical/redundant PCR products, and this is achieved by removing reads sharing the exact same sequence or 5’ alignment positions.
Additional levels of filtering could also be applied to fit the experimental purpose. For example, potential undigested restriction sites could be specifically filtered out, rather than passively identified, by removing reads mapped to the same chromosomal strand with a small distance (user-defined, experience-based) in between.

Binning and Bin-Level Filtering
Based on their midpoint coordinates, Hi-C restriction fragments are binned into fixed genomic intervals, with bin sizes ranging from 40 kb to 1 Mb.
The rationale behind this approach is that by reducing the complexity of the data and lowering the number of candidate genome-wide interactions per bin, genomic bins allow for the construction of more robust and less noisy signals, in the form of contact frequencies, at the expense of resolution (though restriction fragment length still remains the ultimate physical limit to Hi-C resolution).
Bin to bin interactions are aggregated by simply taking the sum, although more focused and informative methods have also been developed over the years to further enhance the signal.
One such method described by Rao et al. aims to push the limit of bin size to smaller and smaller bins, eventually having > 80% of bins covered by 1000 reads each, which significantly increased the resolution of the final analysis results.
Bin-level filtering, just like fragment-level filtering, also takes place to shed experimental artifacts from the obtained data. Bins with high noise and low signals are removed as they typically represent highly repetitive genomic contents around the
telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes. Although there are different architectures, telomeres, in a broad sense, are a widespread genetic feature mo ...
s and
centromere
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers ...
s.
This is done by comparing the individual bin sums to the sum of all bins and removing the bottom 1% of bins, or by using the variance as a measure of noise.
Low-coverage bins, or bins three standard deviations below the center of a log-normal distribution (which fits the total number of contacts per genomic bin), are removed using the MAD-max (maximum allowed median absolute deviation) filter.
After binning, Hi-C data will be stored in a symmetrical matrix format.
More recently, many approaches have been proposed to predetermine the optimal bin size for different Hi-C experiments. Li et al. in 2018 described deDoc, a method where bin size is selected as the one at which the structural entropy of the Hi-C matrix reaches a stable minimum.
QuASAR, on the other hand, offers a bit more quality assessment, and compares replicate scores of the samples (given that replicates are indeed included for the experimental purpose) to find the maximum usable resolution.
Some publications
also tried to score interaction frequencies at the single-fragment level, where a higher coverage can be achieved even with a lower number of reads. HiCPlus,
a tool developed by Zhang et al. in 2018, is able to impute Hi-C matrices similar to the original ones using only 1/16 of the original reads.
Balancing/normalization
Balancing refers to the process of bias correction of the obtained Hi-C data, and can be either explicit or implicit.
Explicit balancing methods require the explicit definitions of biases known to be associated with Hi-C reads (or any
high-throughput sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The ...
technique in general) including the read mappability,
GC content
In molecular biology and genetics, GC-content (or guanine-cytosine content) is the percentage of nitrogenous bases in a DNA or RNA molecule that are either guanine (G) or cytosine (C). This measure indicates the proportion of G and C bases out of ...
, as well as individual fragment length.
A correction factor is first computed for each of the considered biases, followed by each of their combination, and then applied to the read counts per genomic bin.
However, some biases can come from an unknown origin, in which case an implicit balancing approach is used instead. Implicit balancing relies on the assumption that each genomic locus should have “equal visibility”, which suggests that the interaction signal at each genomic locus in the Hi-C data should add up to the same total amount.
One approach called iterative correction uses the
Sinkhorn–Knopp balancing algorithm and attempts to balance the symmetrical matrix using the aforementioned assumption (by equalizing the sum of each and every row and column in the matrix).
The algorithm iteratively alternates between two steps: 1) dividing each row by its mean, and 2) dividing each column by its mean, which are guaranteed to converge in the end and leave no obviously high rows or columns in the interaction matrix.
Other computational methods also exist to normalize the biases inherent to Hi-C data, including sequential component normalization (SCN),
the Knight-Ruiz matrix-balancing approach,
and eigenvector decomposition (ICE) normalization.
In the end, both the explicit and the implicit bias correction methods yield comparable results.
Analysis and Data Interpretation
With a binned, genome-wide interaction matrix, common interaction patterns observed in mammalian genomes can be identified and interpreted biologically, while more rare, less frequently observed patterns such as
circular chromosome
A circular chromosome is a chromosome in bacteria, archaea, mitochondria, and chloroplasts, in the form of a molecule of circular DNA, unlike the linear chromosome of most eukaryotes.
Most prokaryote chromosomes contain a circular DNA molecul ...
s and centromere clustering, may require additional specially-tailored methods to be identified.
1. Cis/Trans Interaction Ratio
Cis
Cis or cis- may refer to:
Places
* Cis, Trentino, in Italy
* In Poland:
** Cis, Świętokrzyskie Voivodeship, south-central
** Cis, Warmian-Masurian Voivodeship, north
Math, science and biology
* cis (mathematics) (cis(''θ'')), a trigonome ...
/
trans interactions are one of the two strongest interaction patterns observed in Hi-C maps.
They are not locus-specific, and thus are considered as a genome-level pattern.
Typically, a higher interaction frequency is observed, on average, for pairs of loci residing on the same chromosome (in cis) than pairs of loci residing on different chromosomes (in trans).
In Hi-C interaction matrices, cis/trans interactions appear as square blocks centered along a diagonal, matching individual chromosomes at the same time.
Because this pattern is relatively consistent across different species and cell types, it can be used to assess the quality of the data. A noisier experiment, due to random background ligation or any unknown factor, will result in a lower cis to trans interaction ratio (as the noise is expected to affect both cis and trans interactions to a similar extent), and high-quality experiments typically have a cis/trans interaction ratio between 40 and 60 for the human genome.
2. Distance-dependent interaction frequency
This pattern refers to the distance-dependent decay of interaction frequencies on a genome level, and represents the second one of the two strongest Hi-C interaction patterns.
As the interaction frequencies between cis-interacting loci decrease (as a result of further distance between them), a gradual decrease of interaction frequency can be observed moving away from the diagonal in the interaction matrix.
Various polymer models
exist to statistically characterize the properties of loci pairs separated by a given distance, but discrete binning and fitting continuous functions are two common ways to analyze the distance-dependent interaction frequencies between datapoints.
First, interaction frequencies can be binned based on their genomic distance, then a continuous function is fitted to the data using information of the average of each bin.
The resulting decay function is plotted on a log-log plot so that a linear line can be used to represent the power-law decays predicted by polymer models.
However, often times a simple polymer model will not be sufficient to fully represent the distance-dependent interaction frequencies, at which point more complicated decay functions might result, which might affect the reproducibility of the data due to the presence of locus-specific rather than genome-wide patterns observed in the Hi-C matrix (which are not taken into consideration by polymer models).
3. Chromatin Compartments
The strongest locus-specific pattern found in Hi-C maps is
chromatin compartments,
which takes the shape of a plaid or “checker-board”-like pattern on the interaction matrix, with alternating blocks that range between 1 and 10 Mb in size (which makes them easy to extract even in experiments with very low sampling) in the human genome.
This pattern can be found at both high and low frequencies. Because chromosomes consist of two types of genomic regions that alternate along the length of individual chromosomes, the interaction frequencies between two regions of the same type and interaction frequencies between two regions of different types can be quite different.
The definition of the active (A) and inactive (B) chromatin compartments is based on
principal component analysis
Principal component analysis (PCA) is a popular technique for analyzing large datasets containing a high number of dimensions/features per observation, increasing the interpretability of data while preserving the maximum amount of information, and ...
, first established by Lieberman-Aiden et al. in 2009.
Their approach calculated the correlation of the Hi-C matrix of observed vs. expected signal (obtained from a distance-normalized contact matrix) ratio, and used the sign of the first
eigenvector
In linear algebra, an eigenvector () or characteristic vector of a linear transformation is a nonzero vector that changes at most by a scalar factor when that linear transformation is applied to it. The corresponding eigenvalue, often denote ...
to denote positive and negative parts of the resulting plot as A and B compartments, respectively.
Many genomic studies have indicated that
chromatin compartments are correlated with chromatin states, such as
gene density In genetics, the gene density of an organism's genome is the ratio of the number of genes per number of base pairs, usually written in terms of a million base pairs, or ''megabase'' (Mb). The human genome has a gene density of 11-15 genes/Mb, while ...
, DNA accessibility, GC content,
replication timing, and
histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn ar ...
marks.
Therefore, type A compartments are more specifically defined to represent the gene-dense regions of
euchromatin
Euchromatin (also called "open chromatin") is a lightly packed form of chromatin (DNA, RNA, and protein) that is enriched in genes, and is often (but not always) under active Transcription (genetics), transcription. Euchromatin stands in contrast ...
, while type B compartments represent
heterochromatic
Heterochromia is a variation in coloration. The term is most often used to describe color differences of the iris, but can also be applied to color variation of hair or skin. Heterochromia is determined by the production, delivery, and concent ...
regions with less gene activities.
Overall,
chromatin compartments offer insights on the general organization principles of the genome of interest.
More and more bioinformatics tools capable of performing compartment calling have been developed over the past decade, including HOMER,
HiTC R,
and CscoreTool.
Although they each has their own differences and optimizations made on the original 2009 approach, their base protocols still rely on principal component analysis.
4. Topologically Associating Domains (TADs)
TADs are sub-Mb structures that may harbor gene-regulatory features, such as local
promoter
Promoter or Promotor may refer to:
Art, entertainment, and media
* ''The Promoter'' (1952), also known as ''The Card''
* ''The Promoter'' (2012 film)
Professions
* Promoter (entertainment), one who makes arrangements for events or markets them ...
-
enhancer interactions.
More generally, TADs are considered as an emergent property of underlying biological mechanisms, which defines TADs as loop extrusions, compartmentalizations, or any dynamic genomic pattern rather than a static structural feature of the genome.
Thus, TADs represent regulatory microenvironments and usually show up on a Hi-C map as blocks of highly self-interacting regions in which interaction frequencies within the region are significantly higher than interaction frequencies between two adjacent regions.
In Hi-C interaction matrices, TADs are square blocks of elevated interaction frequencies centred along the diagonal.
However, this is merely an oversimplified description, and identifying the actual pattern requires much more statistical processing and estimation.
One approach to identify TADs was described by Dixon et al.,
where they first calculated (within some genomic range) the difference between the average upstream interactions and the average downstream interactions of each bin in the matrix.
This difference was then transformed into a chi-squared statistic based on the Hidden Markov Model, and any sharp change in this chi-squared value, called the directionality index, will define the boundaries of TADs.
Alternatively, one could simply take the ratio between average upstream and downstream interactions to define TAD boundaries, as did Naumova et al.
Another approach is to calculate the average interaction frequencies crossing over each bin, again within some predetermined genomic range.
The resulting value is referred to as the insulation score and can be thought of as the average of a square sliding along the diagonal of the matrix (Crane et al.).
This value is expected to be lower at TAD boundaries; thus, one can use standard statistical techniques to find
local minima
In mathematical analysis, the maxima and minima (the respective plurals of maximum and minimum) of a function, known collectively as extrema (the plural of extremum), are the largest and smallest value of the function, either within a given ...
(boundaries), and define regions between consecutive boundaries to be TADs.
However, as is increasingly recognized today, TADs represent a hierarchical series of structures that cannot be fully characterized by one-dimensional scores given by the previous methods.
The increased resolution available in newer datasets can now explicitly address TADs with multiscale analysis approaches. As first introduced by Armatus,
resolution specific domains can be identified and a consensus set of domains conserved across resolutions can be calculated,
which transforms the problem of TAD calling into the optimization of scoring functions based on their local interaction densities.
Variations of this approach with different objective functions, such as Lavaburst,
MrTADFinder,
3DNetMod,
and Matryoshka,
are also developed to achieve better computing performance on higher resolution datasets.

5. Point Interactions
Biologically, regulatory interactions usually occur at much smaller scale than TADs, and two genomic elements can activate/inhibit the expression of a gene within as small a distance as 1 kb.
Therefore, point interactions are important in interpreting Hi-C maps, and are expected to appear as local enrichments in contact probability.
However, current methodologies for the identification of point interactions are all implicit in nature, in that they do not instruct what a point interaction should look like.
Instead, point mutations are identified as
outlier
In statistics, an outlier is a data point that differs significantly from other observations. An outlier may be due to a variability in the measurement, an indication of novel data, or it may be the result of experimental error; the latter are ...
s with higher interaction frequencies than expected within the Hi-C matrix, given that the background model consists only of the strongest signals such as the distance-decay functions.
The background model can be estimated and constructed using both local signal distributions and global approaches (i.e. chromosome-wide/genome-wide).
Many of the aforementioned bioinformatics packages incorporate algorithms to identify point interactions. In short, the significance of individual pairwise interaction is calculated, and significantly high outliers are corrected for multiple testing before they are recognized as truly informative point interactions.
It is helpful to compliment identified point interactions with additional evidence such as analysis of enrichment scores and biological replicates, to indicate that these interactions are indeed of biological significance.
Uses
Development
1. Cell Division
Hi-C can reveal chromatin conformation changes during cell division. In
interphase
Interphase is the portion of the cell cycle that is not accompanied by visible changes under the microscope, and includes the G1, S and G2 phases. During interphase, the cell grows (G1), replicates its DNA (S) and prepares for mitosis (G2). A ...
, chromatins are generally loose and vivacious so that transcription regulation and other regulatory activities could take place.
When entering
mitosis
In cell biology, mitosis () is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis gives rise to genetically identical cells in which the total number of chromosomes is maint ...
and cell division, chromatins become compactly folded into dense cylindrical chromosomes.
Within the past five years, the development of single-cell Hi-C has enabled the depiction of the entire 3D structural landscape of chromatins/chromosomes throughout the
cell cycle
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA ( DNA replication) and some of its organelles, and sub ...
, and many studies have discovered that these identified genomic domains remain unchanged in interphase, and are erased by silencing mechanisms when the cell enters mitosis.
When mitotic division is completed and the cell re-enters the interphase, chromatin 3D structures are observed to be re-established, and transcription regulation is restored.
2. Transcription Regulation and Fate Determination
It has been suspected that the differentiation of
embryonic stem cells (ESCs) and
induced pluripotent stem cells (iPSCs) into various mature
cell lineage
Cell lineage denotes the developmental history of a tissue or organ from the fertilized embryo. This is based on the tracking of an organism's cellular ancestry due to the cell divisions and relocation as time progresses, this starts with the orig ...
s is accompanied by global changes in chromosomal structures and consequently interaction dynamics to allow for the regulation of transcriptional activation/silencing.
Standard Hi-C can be used to investigate this research question.
In 2015, Dixon et al.
applied standard Hi-C to capture global 3D dynamics in human ESCs during their differentiation into
high five cells High Five (BTI-Tn-5B1-4) is an insect cell line that originated from the ovarian cells of the cabbage looper, ''Trichoplusia ni.'' It was developed by the Boyce Thompson Institute for Plant Research.
High Five cells have become one of the most co ...
. Due to the ability of Hi-C to depict dynamic interactions in differentiation-related TADs, the researchers discovered increases in the number of DHS sites,
CTCF
Transcriptional repressor CTCF also known as 11-zinc finger protein or CCCTC-binding factor is a transcription factor that in humans is encoded by the ''CTCF'' gene. CTCF is involved in many cellular processes, including transcriptional regulatio ...
binding ability,
active histone modifications, and target gene expressions within these TADs of interest, and found significant participation of major
pluripotency Pluripotency: These are the cells that can generate into any of the three Germ layers which imply Endodermal, Mesodermal, and Ectodermal cells except tissues like the placenta.
According to Latin terms, Pluripotentia means the ability for many thin ...
factors such as
OCT4
Oct-4 ( octamer-binding transcription factor 4), also known as POU5F1 ( POU domain, class 5, transcription factor 1), is a protein that in humans is encoded by the ''POU5F1'' gene. Oct-4 is a homeodomain transcription factor of the POU family. ...
, NANOG, and
SOX2 in the interaction network during
somatic cell
A somatic cell (from Ancient Greek σῶμα ''sôma'', meaning "body"), or vegetal cell, is any biological cell forming the body of a multicellular organism other than a gamete, germ cell, gametocyte or undifferentiated stem cell. Such cells co ...
reprogramming.
Since then, Hi-C has been recognized as one of the standard methods to probe for transcriptional regulatory activities, and has confirmed that chromosome architecture is closely related to cell fate.
3. Growth and Development
Mammalian somatic growth and development starts with the
fertilization
Fertilisation or fertilization (see spelling differences), also known as generative fertilisation, syngamy and impregnation, is the fusion of gametes to give rise to a new individual organism or offspring and initiate its development. Pro ...
of
sperm and
oocyte
An oocyte (, ), oöcyte, or ovocyte is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in a female fetus in the ovary during female gametogenesis. The femal ...
, followed by the
zygote
A zygote (, ) is a eukaryotic cell formed by a fertilization event between two gametes. The zygote's genome is a combination of the DNA in each gamete, and contains all of the genetic information of a new individual organism.
In multicell ...
stage, the 2-cell, 4-cell, and the 8-cell stage, the
blastocyst
The blastocyst is a structure formed in the early embryonic development of mammals. It possesses an inner cell mass (ICM) also known as the ''embryoblast'' which subsequently forms the embryo, and an outer layer of trophoblast cells called th ...
stage, and finally the
embryo
An embryo is an initial stage of development of a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male sperm ...
stage.
Hi-C made it possible to explore the comprehensive genomic architecture during growth and development, as both sis-Hi-C
and in situ Hi-C
have reported that TADs and genomic A and B compartments are not obviously present and appear to be less well-structured in oocyte cells.
These structural features of the chromatin only gradually establish from weaker frequencies to cleaner and more frequent datapoints after fertilization, as developmental stages progress.
Genome Evolution
As data on 3D genome structures becomes more and more prevalent in recent years, Hi-C begins to be used as a means to track evolutionary structural features/changes. Genomic
single nucleotide polymorphisms (SNPs) and TADs are typically conserved across species,
along with the CTCF factor in the chromatin domain evolution.
Other factors, however, have been revealed by Hi-C techniques to experience structural evolutions in 3D architecture. These include
codon usage
Codon usage bias refers to differences in the frequency of occurrence of synonymous codons in coding DNA. A codon is a series of three nucleotides (a triplet) that encodes a specific amino acid residue in a polypeptide chain or for the termination ...
frequency similarity (CUFS),
paralog gene co-regulation,
and spatially co-evolving orthologous modules (SCOMs).
For large-scale domain evolution,
chromosomal translocation
In genetics, chromosome translocation is a phenomenon that results in unusual rearrangement of chromosomes. This includes balanced and unbalanced translocation, with two main types: reciprocal-, and Robertsonian translocation. Reciprocal translo ...
s, syntenic regions, as well as genomic rearrangement regions were all relatively conserved.
These findings imply that Hi-C technologies is capable of providing an alternative point of view in the
eukaryotic
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bact ...
tree of life.
Cancer
Several studies have employed the use of Hi-C to describe and study chromatin architecture in different cancers and their impact on disease pathogenesis. Kloetgen et al. used in situ Hi-C to study
T cell acute lymphoblastic leukemia (T-ALL) and found a TAD fusion event that removed a CTCF insulation site, allowing for the oncogene
MYC
''Myc'' is a family of regulator genes and proto-oncogenes that code for transcription factors. The ''Myc'' family consists of three related human genes: ''c-myc'' ( MYC), ''l-myc'' ( MYCL), and ''n-myc'' ( MYCN). ''c-myc'' (also sometimes ref ...
’s promoter to directly interact with a distal
super enhancer.
Fang et al. have also shown how there are T-ALL specific gain or loss of chromatin insulation, which alters the strength of TAD architecture of the genome, using in situ Hi-C.
[{{cite journal , last1=Fang , first1=Celestia , last2=Wang , first2=Zhenjia , last3=Han , first3=Cuijuan , last4=Safgren , first4=Stephanie L. , last5=Helmin , first5=Kathryn A. , last6=Adelman , first6=Emmalee R. , last7=Serafin , first7=Valentina , last8=Basso , first8=Giuseppe , last9=Eagen , first9=Kyle P. , last10=Gaspar-Maia , first10=Alexandre , last11=Figueroa , first11=Maria E. , last12=Singer , first12=Benjamin D. , last13=Ratan , first13=Aakrosh , last14=Ntziachristos , first14=Panagiotis , last15=Zang , first15=Chongzhi , title=Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation , journal=Genome Biology , date=15 September 2020 , volume=21 , issue=1 , pages=247 , doi=10.1186/s13059-020-02152-7 , pmid=32933554 , pmc=7493976 , issn=1474-760X] Low-C has been used to map the chromatin structure of primary
B cell
B cells, also known as B lymphocytes, are a type of white blood cell of the lymphocyte subtype. They function in the humoral immunity component of the adaptive immune system. B cells produce antibody molecules which may be either secreted or ...
s of a
diffuse large B-cell lymphoma
Diffuse large B-cell lymphoma (DLBCL) is a cancer of B cells, a type of lymphocyte that is responsible for producing antibodies. It is the most common form of non-Hodgkin lymphoma among adults, with an annual incidence of 7–8 cases per 100,000 ...
patient and was used to find high chromosome structural variation between the patient and healthy B-cells.
Overall, the application of Hi-C and its variants in cancer research provides unique insight into the molecular underpinnings of the driving factors of cell abnormality.
It can help explain biological phenomena (high MYC expression in T-ALL) and help aid drug development to target mechanisms unique to cancerous cells.
References
Genomics techniques
 ]
Hi-C (or standard Hi-C) is a high-throughput Genomics, genomic and epigenomic technique first described in 2009 by Lieberman-Aiden et al. to capture chromatin conformation. In general, Hi-C is considered as a derivative of a series of
]
Hi-C (or standard Hi-C) is a high-throughput Genomics, genomic and epigenomic technique first described in 2009 by Lieberman-Aiden et al. to capture chromatin conformation. In general, Hi-C is considered as a derivative of a series of  Cell and nuclear membranes are highly permeable to formaldehyde. Formaldehyde cross-linking is frequently employed for the detection and quantification of DNA-protein and protein-protein interactions. Of interest in the context of Hi-C, and all 3C-based methods, is the ability of formaldehyde to capture cis chromosomal interactions between distal segments of chromatin. It does so by forming covalent links between spatially adjacent chromatin segments. Formaldehyde can react with
Cell and nuclear membranes are highly permeable to formaldehyde. Formaldehyde cross-linking is frequently employed for the detection and quantification of DNA-protein and protein-protein interactions. Of interest in the context of Hi-C, and all 3C-based methods, is the ability of formaldehyde to capture cis chromosomal interactions between distal segments of chromatin. It does so by forming covalent links between spatially adjacent chromatin segments. Formaldehyde can react with 



