Demethylase on:
[Wikipedia]
[Google]
[Amazon]
Demethylases are enzymes that remove methyl (CH3) groups from
 Histone methylation was initially considered an effectively irreversible process as the half-life of the histone methylation was approximately equal to the histone half-life. Histone lysine demethylase LSD1 (later classified as KDM1A) was first identified in 2004 as a nuclear amine oxidase homolog. Two main classes of histone lysine demethylases exist, defined by their mechanisms: flavin adenine dinucleotide (FAD)-dependent amine oxidases and α-ketoglutarate-dependent hydroxylases.
Histone lysine demethylases possess a variety of domains that are responsible for histone recognition, DNA binding, methylated
Histone methylation was initially considered an effectively irreversible process as the half-life of the histone methylation was approximately equal to the histone half-life. Histone lysine demethylase LSD1 (later classified as KDM1A) was first identified in 2004 as a nuclear amine oxidase homolog. Two main classes of histone lysine demethylases exist, defined by their mechanisms: flavin adenine dinucleotide (FAD)-dependent amine oxidases and α-ketoglutarate-dependent hydroxylases.
Histone lysine demethylases possess a variety of domains that are responsible for histone recognition, DNA binding, methylated  ;KDM1:The KDM1 homologs include
;KDM1:The KDM1 homologs include
 Another example of a demethylase is protein-glutamate methylesterase, also known as CheB protein (EC 3.1.1.61), which demethylates MCPs (methyl-accepting chemotaxis proteins) through hydrolysis of carboxylic ester bonds. The association of a chemotaxis receptor with an agonist leads to the phosphorylation of CheB. Phosphorylation of CheB protein enhances its catalytic MCP demethylating activity resulting in adaption of the cell to environmental stimuli. MCPs respond to extracellular attractants and repellents in bacteria like '' E. coli'' in chemotaxis regulation. CheB is more specifically termed a methylesterase, as it removes methyl groups from
Another example of a demethylase is protein-glutamate methylesterase, also known as CheB protein (EC 3.1.1.61), which demethylates MCPs (methyl-accepting chemotaxis proteins) through hydrolysis of carboxylic ester bonds. The association of a chemotaxis receptor with an agonist leads to the phosphorylation of CheB. Phosphorylation of CheB protein enhances its catalytic MCP demethylating activity resulting in adaption of the cell to environmental stimuli. MCPs respond to extracellular attractants and repellents in bacteria like '' E. coli'' in chemotaxis regulation. CheB is more specifically termed a methylesterase, as it removes methyl groups from 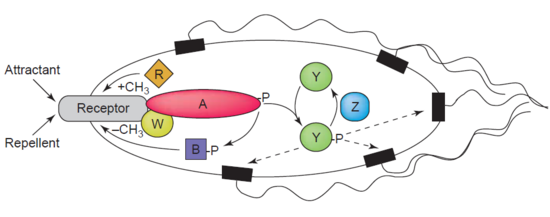
nucleic acid
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main ...
s, proteins (particularly histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn ar ...
s), and other molecules. Demethylases are important epigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are " ...
proteins, as they are responsible for transcriptional regulation
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA ( transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from al ...
of the genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ...
by controlling the methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These t ...
of DNA and histones, and by extension, the chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important ...
state at specific gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
loci.
Histone lysine demethylation
amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha ...
substrate binding and catalytic activity. These include:
* FAD-dependent amine oxidase domains containing the active catalytic site of KDM1
* Jumonji-C domains containing the active catalytic site of KDM2 through KDM8
* Jumonji-N domains responsible for Jumonji-C domain conformation stability
* SWIRM (SWI3P, RSC8P and Moira) domains proposed as an anchor site for histone substrates and responsible for chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important ...
stability
* PHD, CXXC and C5HC2 zinc finger
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized struct ...
domains responsible for histone recognition and binding
Histone lysine demethylases are classified according to their domains and unique substrate specificities. The lysine substrates and identified according to their position in the corresponding histone amino acid sequence and methylation state (for example, H3K9me3 refers to trimethylated histone 3 lysine 9.)  ;KDM1:The KDM1 homologs include
;KDM1:The KDM1 homologs include KDM1A
Lysine-specific histone demethylase 1A (LSD1) also known as lysine (K)-specific demethylase 1A (KDM1A) is a protein in humans that is encoded by the KDM1A gene. LSD1 is a flavin-dependent monoamine oxidase, which can demethylate mono- and di-met ...
and KDM1B. KDM1A demethylates H3K4me1/2 and H3K9me1/2, and KDM1B emethylates H3K4me1/2. KDM1 activity is critical to embryogenesis
An embryo is an initial stage of development of a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male sperm ...
and tissue-specific differentiation, as well as oocyte growth. Deletion of the gene for KDM1A can have effects on the growth and differentiation of embryonic stem cells
Embryonic stem cells (ESCs) are pluripotent stem cells derived from the inner cell mass of a blastocyst, an early-stage pre- implantation embryo. Human embryos reach the blastocyst stage 4–5 days post fertilization, at which time they cons ...
and is universally lethal in knockout mice. KDM1A gene expression is observed to be upregulated in some cancers, and so KDM1A inhibition has therefore been considered a possible epigenetic treatment for cancer.:KDM1B, however, is mostly involved in oocyte
An oocyte (, ), oöcyte, or ovocyte is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in a female fetus in the ovary during female gametogenesis. The femal ...
development. Deletion of this gene leads to maternal effect
A maternal effect is a situation where the phenotype of an organism is determined not only by the environment it experiences and its genotype, but also by the environment and genotype of its mother. In genetics, maternal effects occur when an orga ...
lethality in mice. Orthologs of KDM1 in '' D. melanogaster'' and ''C. elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek ''caeno-'' (recent), ''rhabditis'' ( ...
'' appear to function similarly to KDM1B rather than KDM1A.
;KDM2:The KDM2 homologs include KDM2A and KDM2B. KDM2A and KDM2B demethylate H3K4me3 and H3K36me2/3. KDM2A has roles in either promoting or inhibiting tumor function, and KDM2B has roles in oncogenesis. KDM2A and KDM2B possess CXXC zinc finger domains responsible for binding to unmethylated CpG islands, and it is believed that they may bind to many gene regulatory elements in the absence of sequence-specific transcription factors.:Overexpressed KDM2B has been observed in human lymphoma
Lymphoma is a group of blood and lymph tumors that develop from lymphocytes (a type of white blood cell). In current usage the name usually refers to just the cancerous versions rather than all such tumours. Signs and symptoms may include enl ...
and adenocarcinoma
Adenocarcinoma (; plural adenocarcinomas or adenocarcinomata ) (AC) is a type of cancerous tumor that can occur in several parts of the body. It is defined as neoplasia of epithelial tissue that has glandular origin, glandular characteristics, o ...
, and underexpressed KDM2B has been observed in human prostate cancer and glioblastoma. KDM2B has been additionally shown to prevent senescence
Senescence () or biological aging is the gradual deterioration of functional characteristics in living organisms. The word ''senescence'' can refer to either cellular senescence or to senescence of the whole organism. Organismal senescence inv ...
in some cells through ectopic expression.
;KDM3:The KDM3 homologs include KDM3A
Lysine demethylase 3A is a protein that in humans is encoded by the KDM3A gene.
Function
This gene encodes a zinc finger protein that contains a jumonji C (JmjC) domain and may play a role in hormone-dependent Transcription (biology), transcrip ...
, KDM3B and KDM3C. KDM3A, KDM3B and KDM3C demethylate H3K9me1/2. KDM3A has roles in spermatogenesis
Spermatogenesis is the process by which haploid spermatozoa develop from germ cells in the seminiferous tubules of the testis. This process starts with the mitotic division of the stem cells located close to the basement membrane of the t ...
and metabolic functions, however, the activity of KDM3B and KDM3C are not specifically known.:Knockdown studies of KDM3A in mice resulted in male infertility and adult onset-obesity. Additional studies have indicated that KDM3A may play a role in regulation of androgen receptor-dependent genes as well as genes involved in pluripotency, indicating a potential role for KDM3A in tumorigenesis.
;KDM4:The KDM4 homologs include KDM4A, KDM4B, KDM4C, KDM4D, KDM4E and KDM4F. KDM4A, KDM4B and KDM4C demethylate H3K9me2/3, H3K9me3 and H3K36me2/3, and KDM4D, KDM4E and KDM4F demethylate H3K9me2/3. KDM4A, KDM4B, KDM4C and KDM4D have roles in tumorigenesis
Carcinogenesis, also called oncogenesis or tumorigenesis, is the formation of a cancer, whereby normal cells are transformed into cancer cells. The process is characterized by changes at the cellular, genetic, and epigenetic levels and abn ...
, however, the activity of KDM4E and KDM4F are not specifically known.. KDM4B upregulation has bee observed in medulloblastoma, and KDM4C amplification has been documented in oesophageal squamous carcinoma, medulloblastoma and breast cancer. Other gene expression data has also suggested KDM4A, KDM4B, and KDM4C are overexpressed in prostate cancer.
;KDM5:The KDM5 homologs includes KDM5A, KDM5B
Lysine-specific demethylase 5B also known as histone demethylase JARID1B is a demethylase enzyme that in humans is encoded by the ''KDM5B'' gene. JARID1B belongs to the alpha-ketoglutarate-dependent hydroxylase superfamily.
Function
Jarid1B ...
, KDM5C and KDM5D. KDM5A, KDM5B, KDM5C and KDM5D demethylate H3K4me2/3. The KDM5 family appears to regulate key developmental functions, including cellular differentiation, mitochondrial function and cell cycle
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA ( DNA replication) and some of its organelles, and sub ...
progression. KDM5B and KDM5C have also shown to interaction with PcG proteins, which are involved in transcriptional repression. KDM5C mutations on the X-chromosome have also been observed in patients with X-linked intellectual disability. Depletion of KDM5C homologs in '' D. rerio'' have shown brain-patterning defects and neuronal cell death.
;KDM6:The KDM6 family includes KDM6A, KDM6B and KDM6C. KDM6A and KDM6B demethylate H3K27me2/3, and KDM4C demethylates H3K27me3. KDM6A and KDM6B possess tumor-suppressive characteristics. KDM6A knockdowns in fibroblast
A fibroblast is a type of biological cell that synthesizes the extracellular matrix and collagen, produces the structural framework ( stroma) for animal tissues, and plays a critical role in wound healing. Fibroblasts are the most common cells of ...
s lead to an immediate increase in fibroblast population. KDM6B expressed in fibroblasts induces oncogenes of the Ras/Raf/MEK/ERK pathway. Point mutations of KDM6A have been identified as one cause of Kabuki syndrome
Kabuki syndrome (also previously known as Kabuki-makeup syndrome (KMS) or Niikawa–Kuroki syndrome) is a congenital disorder of genetic origin. It affects multiple parts of the body, with varying symptoms and severity, although the most common is ...
, a congenital disorder resulting in intellectual disability. Deletion of KDM6A in '' D. rerio'' results in decreased expression of HOX genes, which play a role in regulating body patterning during development. In mammalian studies, KDM6A has been shown to regulate HOX genes as well. Mutation of KDM5B disrupt gonad development in ''C.elegans''. Other studies have shown that KDM6B expression is upregulated in activated macrophages
Macrophages (abbreviated as M φ, MΦ or MP) ( el, large eaters, from Greek ''μακρός'' (') = large, ''φαγεῖν'' (') = to eat) are a type of white blood cell of the immune system that engulfs and digests pathogens, such as cancer ce ...
and dynamically expressed during differentiation of stem cells.
Ester demethylation
 Another example of a demethylase is protein-glutamate methylesterase, also known as CheB protein (EC 3.1.1.61), which demethylates MCPs (methyl-accepting chemotaxis proteins) through hydrolysis of carboxylic ester bonds. The association of a chemotaxis receptor with an agonist leads to the phosphorylation of CheB. Phosphorylation of CheB protein enhances its catalytic MCP demethylating activity resulting in adaption of the cell to environmental stimuli. MCPs respond to extracellular attractants and repellents in bacteria like '' E. coli'' in chemotaxis regulation. CheB is more specifically termed a methylesterase, as it removes methyl groups from
Another example of a demethylase is protein-glutamate methylesterase, also known as CheB protein (EC 3.1.1.61), which demethylates MCPs (methyl-accepting chemotaxis proteins) through hydrolysis of carboxylic ester bonds. The association of a chemotaxis receptor with an agonist leads to the phosphorylation of CheB. Phosphorylation of CheB protein enhances its catalytic MCP demethylating activity resulting in adaption of the cell to environmental stimuli. MCPs respond to extracellular attractants and repellents in bacteria like '' E. coli'' in chemotaxis regulation. CheB is more specifically termed a methylesterase, as it removes methyl groups from methylglutamate
''N''-Methyl--glutamic acid (methylglutamate) is a chemical derivative of glutamic acid in which a methyl group has been added to the amino group. It is an intermediate in methane metabolism. Biosynthetically, it is produced from methylamine ...
residues located on the MCPs through hydrolysis, producing glutamate
Glutamic acid (symbol Glu or E; the ionic form is known as glutamate) is an α-amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a non-essential nutrient for humans, meaning that the human body can syn ...
accompanied by the release of methanol.
CheB is of particular interest to researchers as it may be a therapeutic target for mitigating the spread of bacterial infections.

See also
* Chemotaxis *Esterase
An esterase is a hydrolase enzyme that splits esters into an acid and an alcohol in a chemical reaction with water called hydrolysis.
A wide range of different esterases exist that differ in their substrate specificity, their protein structure ...
* Transferase
A transferase is any one of a class of enzymes that catalyse the transfer of specific functional groups (e.g. a methyl or glycosyl group) from one molecule (called the donor) to another (called the acceptor). They are involved in hundreds of ...
* Methylase
References
{{Portal bar, Biology, border=no EC 3.1.1 EC 1.14.11 EC 1.5.8 Signal transduction