Chemical Shift Index on:
[Wikipedia]
[Google]
[Amazon]
 The chemical shift index or CSI is a widely employed technique in
The chemical shift index or CSI is a widely employed technique in
CS23D
ref name=W17> as well as various NMR data analysis web servers such a
RCI
Preditor
ref name=W19> and PANAV have incorporated the CSI method into their software.
CSI calculations via RCI webserver
CSI calculations via Preditor webserver
Chemical shift rereferencing for CSI calculations by Shiftcor
Chemical shift rereferencing for CSI calculations by PANAV
Nuclear magnetic resonance Nuclear magnetic resonance software Protein methods Protein structure Biophysics Scientific techniques
 The chemical shift index or CSI is a widely employed technique in
The chemical shift index or CSI is a widely employed technique in protein nuclear magnetic resonance spectroscopy Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and ...
that can be used to display and identify the location (i.e. start and end) as well as the type of protein secondary structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure ...
(beta strands, helices and random coil regions) found in proteins using only backbone chemical shift data The technique was invented by David S. Wishart
David S. Wishart (born December 7, 1961) is a Canadian researcher and a Distinguished professor, Distinguished University Professor in the Department of Biological Sciences and the Department of Computing Science at the University of Alberta. Wi ...
in 1992 for analyzing 1Hα chemical shifts and then later extended by him in 1994 to incorporate 13C backbone shifts. The original CSI method makes use of the fact that 1Hα chemical shifts of amino acid residues in helices
A helix () is a shape like a corkscrew or spiral staircase. It is a type of smooth space curve with tangent lines at a constant angle to a fixed axis. Helices are important in biology, as the DNA molecule is formed as two intertwined helices, ...
tends to be shifted upfield (i.e. towards the right side of an NMR spectrum) relative to their random coil values and downfield (i.e. towards the left side of an NMR spectrum) in beta strand
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a gen ...
s. Similar kinds of upfield and downfield trends are also detectable in backbone 13C chemical shifts.
Implementation
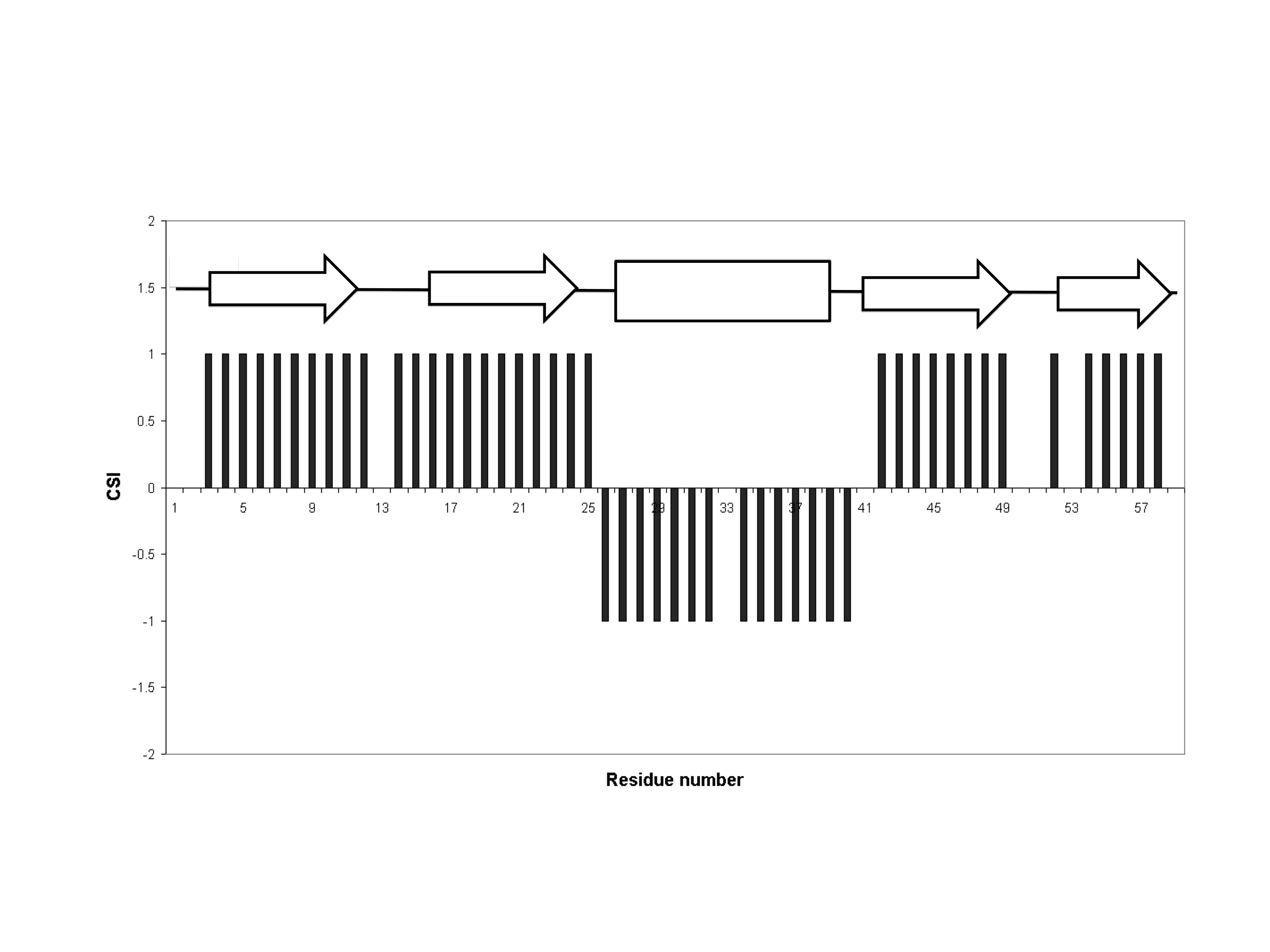
The CSI is a graph-based technique that essentially employs an amino acid-specific digital filter to convert every assigned backbone chemical shift value into a simple three-state (-1, 0, +1) index. This approach generates a more easily understood and much more visually pleasing graph of protein chemical shift values. In particular, if the upfield 1Hα chemical shift (relative to an amino acid-specific random coil value) of a certain residue is > 0.1 ppm, then that amino acid residue is assigned a value of -1. Similarly, if the downfield 1Hα chemical shift of a certain amino acid residue is > 0.1 ppm then that residue is assigned a value of +1. If an amino acid residue's chemical shift is not shifted downfield or upfield by a sufficient amount (i.e. <0.1 ppm), it is given a value of 0. When this 3-state index is plotted as a bar graph over the full length of the protein sequence, simple inspection can allow one to identify beta strands (clusters of +1 values), alpha helices (clusters of -1 values), and random coil segments (clusters of 0 values). A list of the amino acid-specific random coil chemical shifts for CSI calculations is given in Table 1. An example of a CSI graph for a small protein is shown in Figure 1 with the arrows located above the black bars indicating locations of the beta strands and the rectangular box indicating the location of a helix.Performance
Using only 1Hα chemical shifts and simple clustering rules (clusters of 3 or more vertical bars for beta strands and clusters of 4 or more vertical bars for alpha helices), the CSI is typically 75-80% accurate in the identification of secondary structures. This performance depends partly on the quality of the NMR data set as well as the technique (manual or programmatic) used to identify the protein secondary structures. As noted above, a consensus CSI method that filters upfield/downfield chemical shift changes in 13Cα, 13Cβ, and 13C' atoms in a similar manner to 1Hα shifts has also been developed. The consensus CSI combines the CSI plots from backbone 1H and 13C chemical shifts to generate a single CSI plot. It can be up to 85-90% accurate.History
The link between protein chemical shifts and protein secondary structure (specifically alpha helices) was first described by John Markley and colleagues in 1967. With the development of modern 2-dimensional NMR techniques, it became possible to measure more protein chemical shifts. With more peptides and proteins were being assigned in the early 1980s it soon became obvious that amino acid chemical shifts were sensitive not only to helical conformations, but also to β-strand conformations. Specifically, the secondary 1Hα chemical shifts of all amino acids exhibit a clear upfield trend on helix formation and an obvious downfield trend on β-sheet formation. By the early 1990s, a sufficient body of 13C and 15N chemical shift assignments for peptides and proteins had been collected to determine that similar upfield/downfield trends were evident for essentially all backbone 13Cα, 13Cβ, 13C', 1HN and 15N (weakly) chemical shifts. It was these rather striking chemical shift trends that were exploited in the development of the chemical shift index.Limitations
The CSI method is not without some shortcomings. In particular, its performance drops if chemical shift assignments are mis-referenced or incomplete. It is also quite sensitive to the choice of random coil shifts used to calculate the secondary shifts and it generally identifies alpha helices (>85% accuracy) better than beta strands (<75% accuracy) regardless of the choice of random coil shifts. Furthermore, the CSI method does not identify other kinds of secondary structures, such as β-turns. Because of these shortcomings, a number of alternative CSI-like approaches have been proposed. These include: 1) a prediction method that employs statistically derived chemical shift/structure potentials (PECAN); 2) a probabilistic approach to secondary structure identification (PSSI); 3) a method that combines secondary structure predictions from sequence data and chemical shift data (PsiCSI), 4) a secondary structure identification approach that uses pre-specified chemical shift patterns (PLATON) and 5) a two-dimensionalcluster analysis
Cluster analysis or clustering is the task of grouping a set of objects in such a way that objects in the same group (called a cluster) are more similar (in some sense) to each other than to those in other groups (clusters). It is a main task of ...
method known as 2DCSi. The performance of these newer methods is generally slightly better (2-4%) than the original CSI method.
Utility
Since its original description in 1992, the CSI method has been used to characterize the secondary structure of thousands of peptides and proteins. Its popularity is largely due to the fact that it is easy to understand and can be implemented without the need for specialized computer programs. Even though the CSI method can be easily performed manually, a number of commonly used NMR data processing programs such as NMRView, NMR structure generation web servers such aCS23D
ref name=W17> as well as various NMR data analysis web servers such a
RCI
Preditor
ref name=W19> and PANAV have incorporated the CSI method into their software.
See also
* Chemical Shift *Random Coil Index
Random coil index (RCI) predicts protein flexibility by calculating an inverse weighted average of backbone secondary chemical shifts and predicting values of model-free order parameters as well as per-residue RMSD of NMR and molecular dynamics ...
* Protein NMR
* Protein Chemical Shift Re-Referencing
*Protein secondary structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure ...
* Protein Chemical Shift Prediction
* NMR
* Nuclear magnetic resonance spectroscopy
*Protein nuclear magnetic resonance spectroscopy Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and ...
* Protein
References
{{ReflistExternal links
CSI calculations via RCI webserver
CSI calculations via Preditor webserver
Chemical shift rereferencing for CSI calculations by Shiftcor
Chemical shift rereferencing for CSI calculations by PANAV
Nuclear magnetic resonance Nuclear magnetic resonance software Protein methods Protein structure Biophysics Scientific techniques