|
Xenobiology
Xenobiology (XB) is a subfield of synthetic biology, the study of synthesizing and manipulating biological devices and systems. The name "xenobiology" derives from the Greek word ''xenos'', which means "stranger, alien". Xenobiology is a form of biology that is not (yet) familiar to science and is not found in nature. In practice, it describes novel biological systems and biochemistries that differ from the canonical DNA–RNA-20 amino acid system (see central dogma of molecular biology). For example, instead of DNA or RNA, XB explores nucleic acid analogues, termed xeno nucleic acid (XNA) as information carriers. It also focuses on an expanded genetic code and the incorporation of non-proteinogenic amino acids, or “xeno amino acids” into proteins. Difference between xeno-, exo-, and astro-biology "Astro" means "star" and "exo" means "outside". Both exo- and astrobiology deal with the search for naturally evolved life in the Universe, mostly on other planets in the circ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Astrobiology
Astrobiology (also xenology or exobiology) is a scientific field within the List of life sciences, life and environmental sciences that studies the abiogenesis, origins, Protocell, early evolution, distribution, and future of life in the universe by investigating its deterministic conditions and contingent events. As a discipline, astrobiology is founded on the premise that life may exist beyond Earth. Research in astrobiology comprises three main areas: the study of planetary habitability, habitable environments in the Solar System and beyond, the search for planetary biosignatures of past or present extraterrestrial life, and the study of the Abiogenesis, origin and Protocell, early evolution of life on Earth. The field of astrobiology has its origins in the 20th century with the advent of space exploration and the discovery of exoplanets. Early astrobiology research focused on the search for extraterrestrial life and the study of the potential for life to exist on other pl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Nucleic Acid Analogues
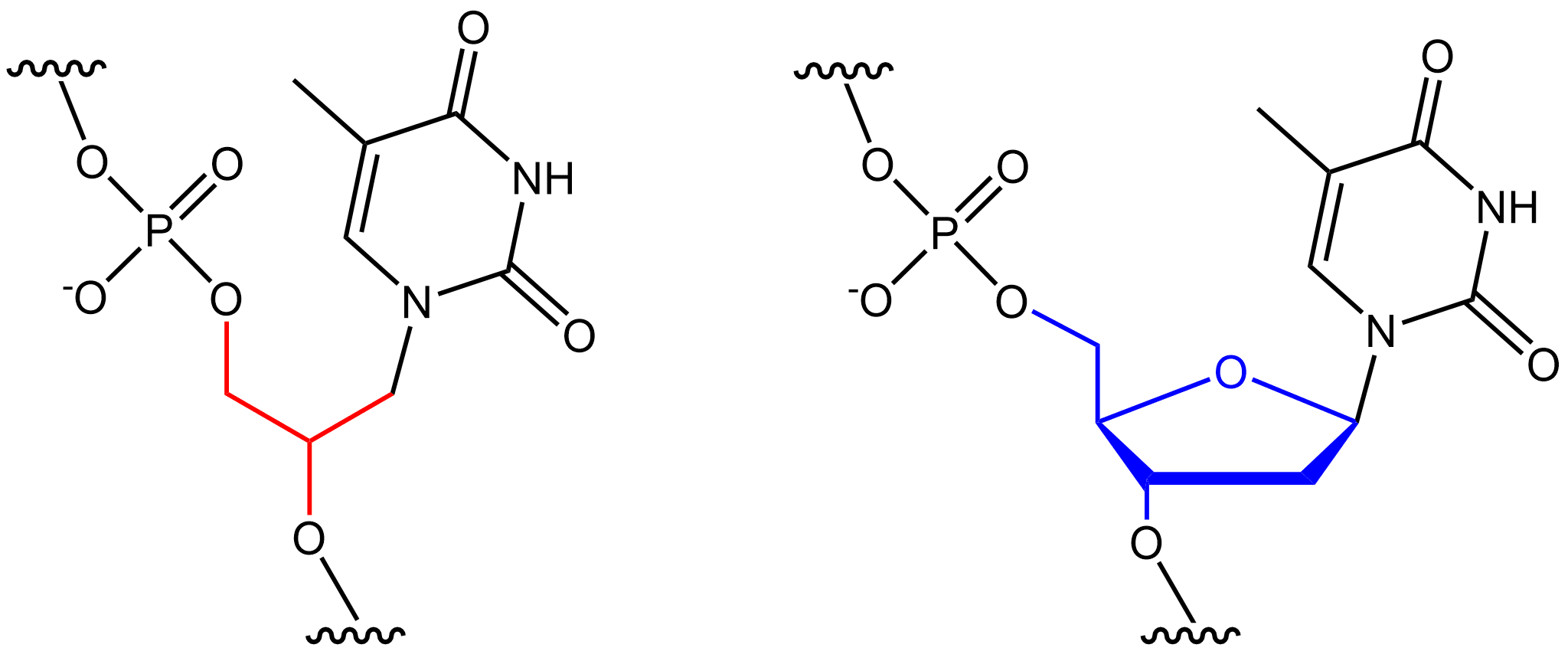
Nucleic acid analogues are compounds which are analogous (structurally similar) to naturally occurring RNA and DNA, used in medicine and in molecular biology research. Nucleic acids are chains of nucleotides, which are composed of three parts: a phosphate backbone, a pentose sugar, either ribose or deoxyribose, and one of four nucleobases. An analogue may have any of these altered. Typically the analogue nucleobases confer, among other things, different base pairing and base stacking properties. Examples include universal bases, which can pair with all four canonical bases, and phosphate-sugar backbone analogues such as PNA, which affect the properties of the chain (PNA can even form a triple helix). Nucleic acid analogues are also called xeno nucleic acids and represent one of the main pillars of xenobiology, the design of new-to-nature forms of life based on alternative biochemistries. Artificial nucleic acids include peptide nucleic acids (PNA), morpholino, and locked nu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Xeno Nucleic Acid
Xenonucleic acids (XNAs) are synthetic nucleic acid analogues that are engineered with structurally distinct components, such as alternative nucleosides, sugars, or backbones. XNAs have fundamentally different properties from endogenous nucleic acids, enabling different specialized applications, such as therapeutics, probes, or functional molecules. For instance, peptide nucleic acids, the backbones of which are made up of repeating aminoethylglycine units, are extremely stable and resistant to degradation by nucleases because they are not recognised. The same nucleobases can be used to store genetic information and interact with DNA, RNA, or other XNA bases, but the different backbone gives the compound different properties. Their altered chemical structure means they cannot be processed by naturally occurring cellular processes. For instance, natural DNA polymerases cannot read and duplicate the alien information, thus the genetic information stored in XNA is invisible to DNA- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Synthetic Biology
Synthetic biology (SynBio) is a multidisciplinary field of science that focuses on living systems and organisms. It applies engineering principles to develop new biological parts, devices, and systems or to redesign existing systems found in nature. It is a branch of science that encompasses a broad range of methodologies from various disciplines, such as biochemistry, biotechnology, biomaterials, Materials science, material science/engineering, genetic engineering, molecular biology, molecular engineering, systems biology, Model lipid bilayer, membrane science, biophysics, Biological engineering, chemical and biological engineering, Electrical engineering, electrical and computer engineering, control engineering and evolutionary biology. It includes designing and constructing BioBrick, biological modules, biological systems, and biological machines, or re-designing existing biological systems for useful purposes. Additionally, it is the branch of science that focuses on the ne ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Expanded Genetic Code
An expanded genetic code is an artificially modified genetic code in which one or more specific codons have been re-allocated to encode an amino acid that is not among the 22 common naturally-encoded proteinogenic amino acids. The key prerequisites to expand the genetic code are: * the non-standard amino acid to encode, * an unused codon to adopt, * a tRNA that recognizes this codon, and * a tRNA synthetase that recognizes only that tRNA and only the non-standard amino acid. Expanding the genetic code is an area of research of synthetic biology, an applied biological discipline whose goal is to engineer living systems for useful purposes. The genetic code expansion enriches the repertoire of useful tools available to science. In May 2019, researchers, in a milestone effort, reported the creation of a new synthetic (possibly artificial) form of viable life, a variant of the bacteria ''Escherichia coli'', by reducing the natural number of 64 codons in the bacterial genome to 61 c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Foldamer
In chemistry, a foldamer is a discrete chain molecule ( oligomer) that folds into a conformationally ordered state in solution. They are artificial molecules that mimic the ability of proteins, nucleic acids, and polysaccharides to fold into well-defined conformations, such as α-helices and β-sheets. The structure of a foldamer is stabilized by noncovalent interactions between nonadjacent monomers. Foldamers are studied with the main goal of designing large molecules with predictable structures. The study of foldamers is related to the themes of molecular self-assembly, molecular recognition, and host–guest chemistry. Design Foldamers can vary in size, but they are defined by the presence of noncovalent, nonadjacent interactions. This definition excludes molecules like poly(isocyanates) (commonly known as polyurethane) and poly(prolines) as they fold into helices reliably due to ''adjacent'' covalent interactions. Foldamers have a dynamic folding reaction (unfold ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
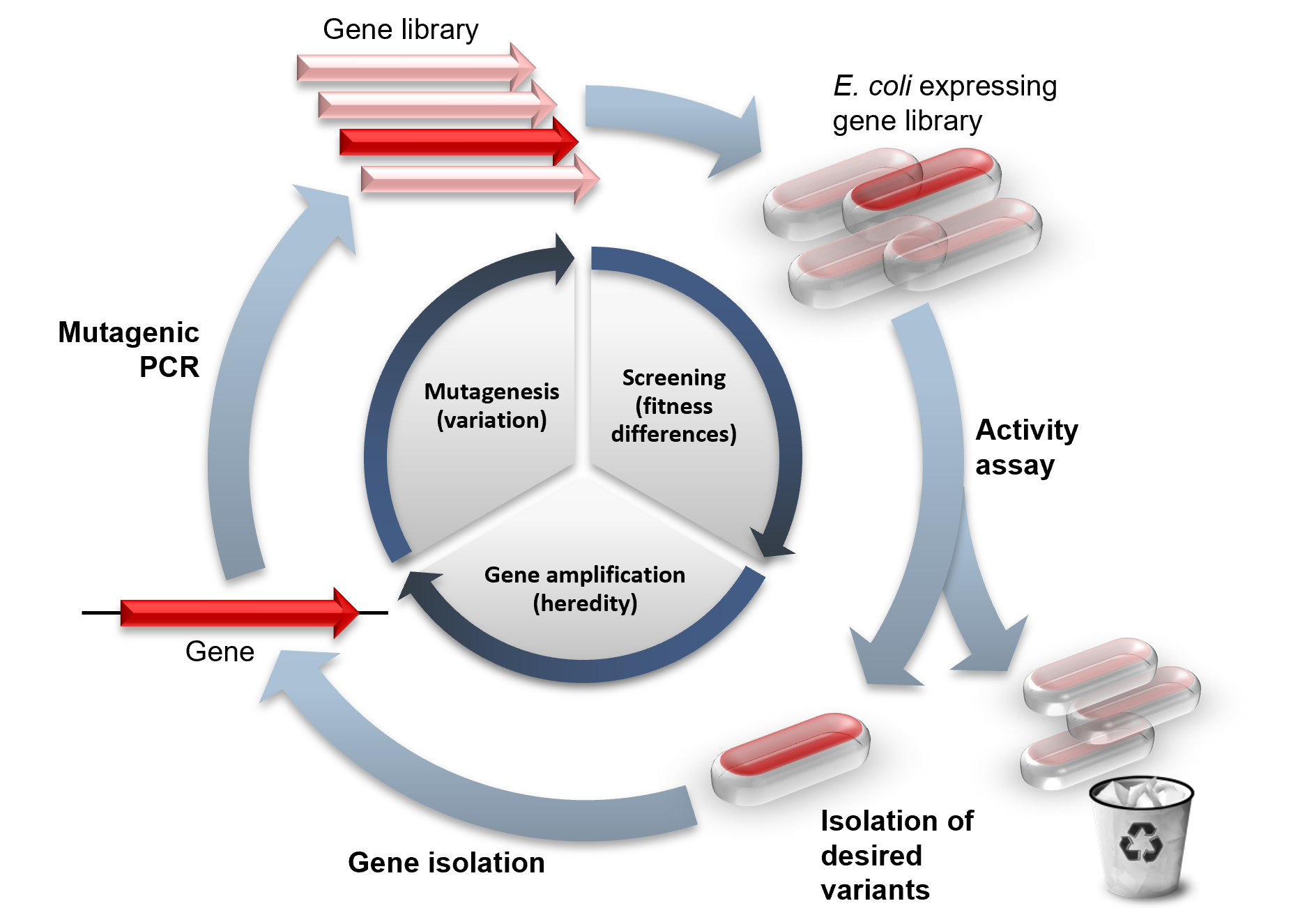
Directed Evolution
Directed evolution (DE) is a method used in protein engineering that mimics the process of natural selection to steer proteins or nucleic acids toward a user-defined goal. It consists of subjecting a gene to iterative rounds of mutagenesis (creating a library of variants), selection (expressing those variants and isolating members with the desired function) and amplification (generating a template for the next round). It can be performed ''in vivo'' (in living organisms), or ''in vitro'' (in cells or free in solution). Directed evolution is used both for protein engineering as an alternative to rationally designing modified proteins, as well as for experimental evolution studies of fundamental evolutionary principles in a controlled, laboratory environment. History Directed evolution has its origins in the 1960s with the evolution of RNA molecules in the "Spiegelman's Monster" experiment. The concept was extended to protein evolution via evolution of bacteria under selec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Body Plan
A body plan, (), or ground plan is a set of morphology (biology), morphological phenotypic trait, features common to many members of a phylum of animals. The vertebrates share one body plan, while invertebrates have many. This term, usually applied to animals, envisages a "blueprint" encompassing aspects such as symmetry (biology), symmetry, Germ layer, layers, segmentation (biology), segmentation, nerve, Limb (anatomy), limb, and Gastrointestinal tract, gut disposition. Evolutionary developmental biology seeks to explain the origins of diverse body plans. Body plans have historically been considered to have evolved in a flash in the Ediacaran biota; filling the Cambrian explosion with the results, and a more nuanced understanding of animal evolution suggests gradual development of body plans throughout the early Palaeozoic. Recent studies in animals and plants started to investigate whether evolutionary constraints on body plan structures can explain the presence of development ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Synthetic Biological Circuit
Synthetic biological circuits are an application of synthetic biology where biological parts inside a Cell (biology), cell are designed to perform logical functions mimicking those observed in electronic circuits. Typically, these circuits are categorized as either genetic circuits, RNA circuits, or protein circuits, depending on the types of biomolecule that interact to create the circuit's behavior. The applications of all three types of circuit range from simply inducing production to adding a measurable element, like green fluorescent protein, to an existing Gene regulatory network, natural biological circuit, to implementing completely new systems of many parts. The goal of synthetic biology is to generate an array of tunable and characterized parts, or modules, with which any desirable synthetic biological circuit can be easily designed and implemented. These circuits can serve as a method to modify cellular functions, create cellular responses to environmental conditions, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Biological Dark Matter
Biological dark matter is an informal term for unclassified or poorly understood genetic material. This genetic material may refer to genetic material produced by unclassified microorganisms. By extension, biological dark matter may also refer to the un-isolated microorganisms whose existence can only be inferred from the genetic material that they produce. Some of the genetic material may not fall under the three existing domains of life: Bacteria, Archaea and Eukaryota; thus, it has been suggested that a possible fourth domain of life may yet be discovered, * although other explanations are also probable. Alternatively, the genetic material may refer to non-coding DNA (so-called "junk DNA") and non-coding RNA produced by known organisms. Genomic dark matter Much of the genomic dark matter is thought to originate from ancient transposable elements and from other low-complexity repetitive elements. Uncategorized genetic material is found in humans and many other species. * Thei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Auxotrophy
Auxotrophy ( "to increase"; ''τροφή'' "nourishment") is the inability of an organism to synthesize a particular organic compound required for its growth (as defined by IUPAC). An auxotroph is an organism that displays this characteristic; ''auxotrophic'' is the corresponding adjective. Auxotrophy is the opposite of prototrophy, which is characterized by the ability to synthesize all the compounds needed for growth. Prototrophic cells are self-sufficient producers of all required metabolites (e.g. amino acids, lipids, cofactors), while auxotrophs require to be on medium with the metabolite that they cannot produce. For example, a methionine auxotrophic cell could only grow on a medium that contained methionine; otherwise, it would starve. In this example, this is because it is unable to produce its own methionine. However, a methionine prototrophic cell would be able to function and replicate on a medium with or without methionine. Replica plating is a technique that transfer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Escherichia Coli
''Escherichia coli'' ( )Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Escherichia'' that is commonly found in the lower intestine of warm-blooded organisms. Most ''E. coli'' strains are part of the normal microbiota of the gut, where they constitute about 0.1%, along with other facultative anaerobes. These bacteria are mostly harmless or even beneficial to humans. For example, some strains of ''E. coli'' benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by harmful pathogenic bacteria. These mutually beneficial relationships between ''E. coli'' and humans are a type of mutualistic biological relationship—where both the humans and the ''E. coli'' are benefitting each other. ''E. coli'' is expelled into the environment within fecal matter. The bacterium grows massi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |