|
Transvection (genetics)
Transvection is an epigenetic phenomenon that results from an interaction between an allele on one chromosome and the corresponding allele on the homologous chromosome. Transvection can lead to either gene activation or repression. It can also occur between nonallelic regions of the genome as well as regions of the genome that are not transcribed. The first observation of mitotic (i.e. non-meiotic) chromosome pairing was discovered via microscopy in 1908 by Nettie Stevens. Edward B. Lewis at Caltech discovered transvection at the '' bithorax'' complex in ''Drosophila'' in the 1950s. Since then, transvection has been observed at a number of additional loci in ''Drosophila'', including the genes known as ''white'', ''decapentaplegic'', ''eyes absent'', ''vestigial'', and ''yellow''. As defined by Lewis, "Operationally, transvection is occurring if the phenotype of a given genotype can be altered solely by disruption of somatic (or meiotic) pairing. Such disruption can generally be acc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Epigenetic
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional (DNA sequence based) genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological traits may result from environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Further, non-coding RNA sequences have been shown to play a key role in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
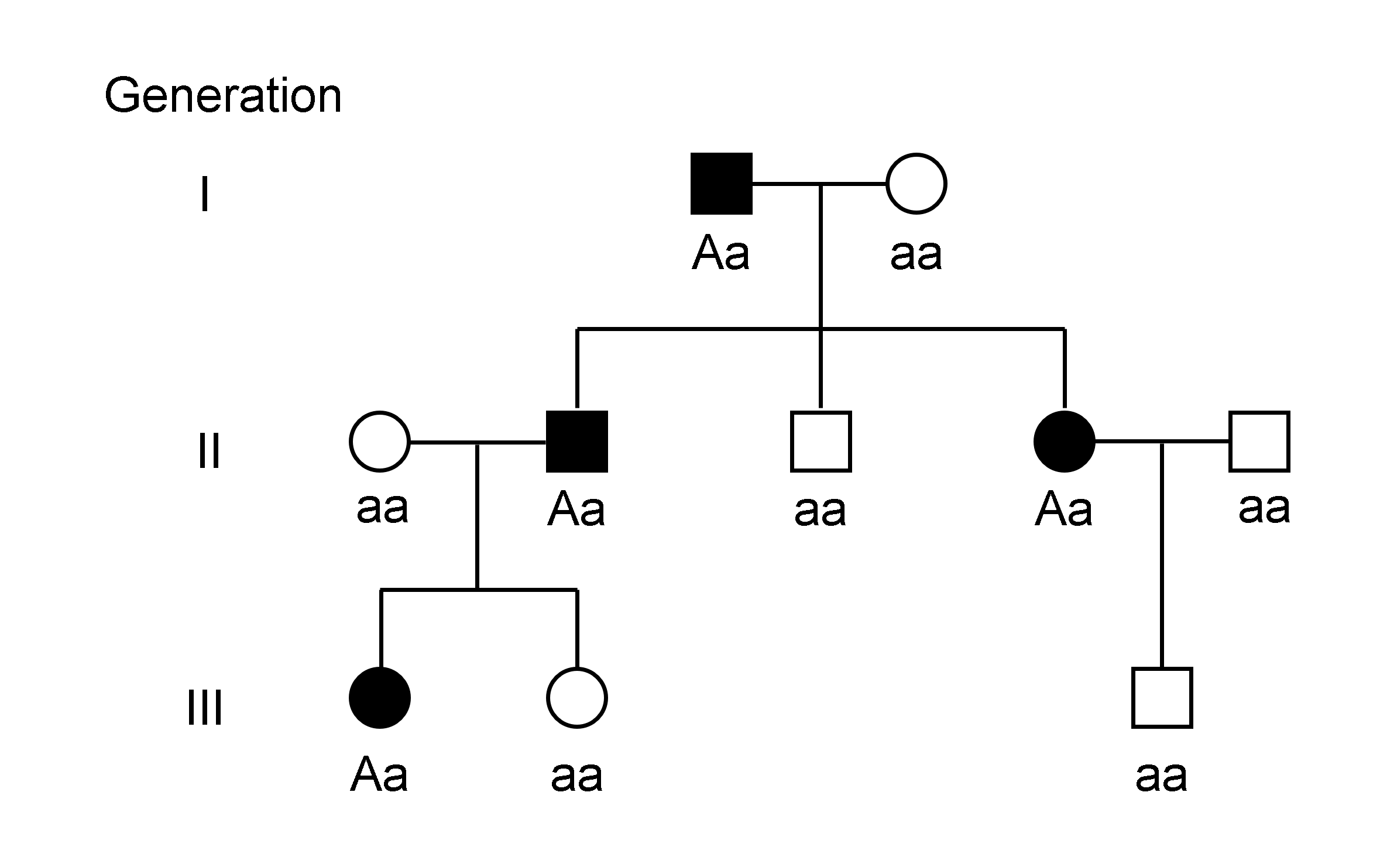
Genotype
The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a specific gene depends on the number of copies of each chromosome found in that species, also referred to as ploidy. In diploid species like humans, two full sets of chromosomes are present, meaning each individual has two alleles for any given gene. If both alleles are the same, the genotype is referred to as Zygosity, homozygous. If the alleles are different, the genotype is referred to as heterozygous. Genotype contributes to phenotype, the observable traits and characteristics in an individual or organism. The degree to which genotype affects phenotype depends on the trait. For example, the petal color in a pea plant is exclusively determined by genotype. The petals can be purple or white depending on the alleles present in the pea plan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Complementation (genetics)
Complementation refers to a genetics, genetic process when two strain (biology), strains of an organism with different homozygous recessive mutations that produce the same mutant phenotype (for example, a change in wing structure in flies) have offspring that express the wild-type phenotype when mated or crossed. Complementation will ordinarily occur if the mutations are in different genes (intergenic complementation). Complementation may also occur if the two mutations are at different sites within the same gene (intragenic complementation), but this effect is usually weaker than that of intergenic complementation. When the mutations are in different genes, each strain's genome supplies the wild-type allele to "complement" the mutated allele of the other strain's genome. Since the mutations are recessive, the offspring will display the wild-type phenotype. A complementation test (sometimes called a "Cistron, cis-trans" test) can test whether the mutations in two strains are in d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Hemizygous
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism. Most eukaryotes have two matching sets of chromosomes; that is, they are diploid. Diploid organisms have the same loci on each of their two sets of homologous chromosomes except that the sequences at these loci may differ between the two chromosomes in a matching pair and that a few chromosomes may be mismatched as part of a chromosomal sex-determination system. If both alleles of a diploid organism are the same, the organism is homozygous at that locus. If they are different, the organism is heterozygous at that locus. If one allele is missing, it is hemizygous, and, if both alleles are missing, it is nullizygous. The DNA sequence of a gene often varies from one individual to another. These gene variants are called alleles. While some gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Physiological Relevance
Physiological relevance is a scientific concept that refers to the applicability or significance of a particular experimental finding or biological observation in the context of normal bodily functions. This concept is often used in biomedical research, where scientists strive to design experiments that not only yield statistically significant results but also have direct implications for understanding human health and disease. Importance in biomedical research Physiological relevance is a critical factor in biomedical research because it helps to bridge the gap between basic science and clinical application. Researchers aim to design studies that not only yield statistically significant results but also have direct implications for understanding human health and disease. For example, a study on the effects of a new drug on cancer cells in a lab dish might show promising results. However, these findings would only be considered physiologically relevant if the drug also demonstrat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Insulator (genetics)
An insulator is a type of cis-regulatory element known as a long-range regulatory element. Found in multicellular eukaryotes and working over distances from the promoter element of the target gene, an insulator is typically 300 bp to 2000 bp in length. Insulators contain clustered binding sites for sequence specific DNA-binding proteins and mediate intra- and inter- chromosomal interactions. Insulators function either as an enhancer-blocker or a barrier, or both. The mechanisms by which an insulator performs these two functions include loop formation and nucleosome modifications. There are many examples of insulators, including the CTCF insulator, the ''gypsy'' insulator, and the β-globin locus. The CTCF insulator is especially important in vertebrates, while the ''gypsy'' insulator is implicated in '' Drosophila.'' The β-globin locus was first studied in chicken and then in humans for its insulator activity, both of which utilize CTCF. The genetic implications of ins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Promotor (biology)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Overview For transcription to take place, the enzyme that synthesizes RNA, known as RNA polymerase, must attach to the DNA near a gene. Promoters contain specific DNA sequences such as response elements that provide a secure initial binding site for RNA polymerase and for proteins called transcription factors that recruit RNA polymerase. These transcription factors hav ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Enhancers
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are ''cis''-acting. They can be located up to 1 Mbp (1,000,000 bp) away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes. Active enhancers typically get transcribed as enhancer or regulatory non-coding RNA, whose expression levels correlate with mRNA levels of target genes. The first discovery of a eukaryotic enhancer was in the immunoglobulin heavy chain gene in 1983. This enhancer, located in the large intron, provided an explanation for the transcriptional activation of rearranged Vh gene promoters while unrearranged Vh promoters remained inactive. Lately, enhancers have been shown to be in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Complementation (genetics)
Complementation refers to a genetics, genetic process when two strain (biology), strains of an organism with different homozygous recessive mutations that produce the same mutant phenotype (for example, a change in wing structure in flies) have offspring that express the wild-type phenotype when mated or crossed. Complementation will ordinarily occur if the mutations are in different genes (intergenic complementation). Complementation may also occur if the two mutations are at different sites within the same gene (intragenic complementation), but this effect is usually weaker than that of intergenic complementation. When the mutations are in different genes, each strain's genome supplies the wild-type allele to "complement" the mutated allele of the other strain's genome. Since the mutations are recessive, the offspring will display the wild-type phenotype. A complementation test (sometimes called a "Cistron, cis-trans" test) can test whether the mutations in two strains are in d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Chromosomal Translocation
In genetics, chromosome translocation is a phenomenon that results in unusual rearrangement of chromosomes. This includes "balanced" and "unbalanced" translocation, with three main types: "reciprocal", "nonreciprocal" and "Robertsonian" translocation. Reciprocal translocation is a chromosome abnormality caused by exchange of parts between non-homologous chromosomes. Two detached fragments of two different chromosomes are switched. Robertsonian translocation occurs when two non-homologous chromosomes get attached, meaning that given two healthy pairs of chromosomes, one of each pair "sticks" and blends together homogeneously. Each type of chromosomal translocation can result in disorders for growth, function and the development of an individuals body, often resulting from a change in their genome. A gene fusion may be created when the translocation joins two otherwise-separated genes. It is detected on cytogenetics or a karyotype of affected cells. Translocations can be bala ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Gene Regulation
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network. Gene regulation is essential for viruses, prokaryotes and eukaryotes as it increases the versatility and adaptability of an organism by allowing the cell to express protein when needed. Although as early as 1951, Barbara McClintock showed interaction between two genetic loci, Activator (''Ac'') and Dissociator (''Ds''), in the color formation of maize seeds, t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Ting Wu
Chao-ting Wu (; born January 24, 1954) is an American molecular biologist. After training at Harvard Medical School in genetics with William Gelbart, at Stanford Medical School with David Hogness, and in a fellowship at Massachusetts General Hospital in molecular biology, Wu began her independent academic career as an assistant professor in anatomy and cellular biology and then genetics at Harvard Medical School in 1993. After a period as a professor of pediatrics in the division of molecular medicine at the Boston Children's Hospital, she returned to the Department of Genetics at Harvard Medical School as a full professor in 2007. Wu's research has focused on the role of chromosome behavior gene activity and inheritance, with emphasis on widespread homology effects, phenomena in which homology between chromosomes plays a role. Her studies have explored transvection in genetics, polycomb-group genes, chromatin pairing and remodeling, and the mechanisms of bridging promoter and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |