|
Telomerase Reverse Transcriptase
Telomerase reverse transcriptase (abbreviated to TERT, or hTERT in humans) is a catalytic subunit of the enzyme telomerase, which, together with the telomerase RNA component (TERC), comprises the most important unit of the telomerase complex. Telomerases are part of a distinct subgroup of RNA-dependent polymerases. Telomerase lengthens telomeres in DNA strands, thereby allowing senescent cells that would otherwise become postmitotic and undergo apoptosis to exceed the Hayflick limit and become potentially immortal, as is often the case with cancerous cells. To be specific, TERT is responsible for catalyzing the addition of nucleotides in a TTAGGG sequence to the ends of a chromosome's telomeres. This addition of repetitive DNA sequences prevents degradation of the chromosomal ends following multiple rounds of replication. hTERT absence (usually as a result of a chromosomal mutation) is associated with the disorder Cri du chat. Function Telomerase is a ribonucleopr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in Cell (biology), cells, by internal metabolism, metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for Transcription (biology), transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MEN1
Menin is a protein that in humans is encoded by the ''MEN1'' gene. Menin is a putative tumor suppressor associated with multiple endocrine neoplasia type 1 (MEN-1 syndrome) and has autosomal dominant inheritance. Variations in the MEN1 gene can cause pituitary adenomas, hyperparathyroidism, pancreatic neuroendocrine tumors, gastrinoma, and adrenocortical cancers. ''In vitro'' studies have shown that menin is localized to the nucleus, possesses two functional nuclear localization signals, and inhibits transcriptional activation by JunD. However, the function of this protein is not known. Two messages have been detected on northern blots but the larger message has not been characterized. Two variants of the shorter transcript have been identified where alternative splicing affects the coding sequence. Five variants where alternative splicing takes place in the 5' UTR have also been identified. History In 1988, researchers at Uppsala University Hospital and the Karolinska I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activating Protein 2
Activating Protein 2 (AP-2) is a family of closely related transcription factor In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...s which plays a critical role in regulating gene expression during early development. References External links * Gene expression Transcription factors {{gene-20-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hypoxia-inducible Factors
Hypoxia-inducible factors (HIFs) are transcription factors that respond to decreases in available oxygen in the cellular environment, or hypoxia. They also respond to instances of pseudohypoxia, such as thiamine deficiency. Both hypoxia and pseudohypoxia leads to impairment of adenosine triphosphate (ATP) production by the mitochondria. Discovery The HIF transcriptional complex was discovered in 1995 by Gregg L. Semenza and postdoctoral fellow Guang Wang. In 2016, William Kaelin Jr., Peter J. Ratcliffe and Gregg L. Semenza were presented the Lasker Award for their work in elucidating the role of HIF-1 in oxygen sensing and its role in surviving low oxygen conditions. In 2019, the same three individuals were jointly awarded the Nobel Prize in Physiology or Medicine for their work in elucidating how HIF senses and adapts cellular response to oxygen availability. Structure Oxygen-breathing species express the highly conserved transcriptional complex HIF-1, which is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sp1 Transcription Factor
Transcription factor Sp1, also known as specificity protein 1* is a protein that in humans is encoded by the ''SP1'' gene. Function The protein encoded by this gene is a zinc finger transcription factor that binds to GC-rich motifs of many promoters. The encoded protein is involved in many cellular processes, including cell differentiation, cell growth, apoptosis, immune responses, response to DNA damage, and chromatin remodeling. post-translational modifications such as phosphorylation, acetylation, ''O''-GlcNAcylation, and proteolytic processing significantly affect the activity of this protein, which can be an activator or a repressor. In the SV40 virus, Sp1 binds to the GC boxes in the regulatory sequence of the genome. Structure SP1 belongs to the Sp/KLF family of transcription factors. The protein is 785 amino acids long, with a molecular weight of 81 kDa. The SP1 transcription factor contains two glutamine-rich activation domains at its N-terminus that ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-Myc
''Myc'' is a family of regulator genes and proto-oncogenes that code for transcription factors. The ''Myc'' family consists of three related human genes: ''c-myc'' ( MYC), ''l-myc'' ( MYCL), and ''n-myc'' ( MYCN). ''c-myc'' (also sometimes referred to as ''MYC'') was the first gene to be discovered in this family, due to homology with the viral gene ''v-myc''. In cancer, ''c-myc'' is often constitutively (persistently) expressed. This leads to the increased expression of many genes, some of which are involved in cell proliferation, contributing to the formation of cancer. A common human translocation involving ''c-myc'' is critical to the development of most cases of Burkitt lymphoma. Constitutive upregulation of ''Myc'' genes have also been observed in carcinoma of the cervix, colon, breast, lung and stomach. Myc is thus viewed as a promising target for anti-cancer drugs. Unfortunately, Myc possesses several features that have rendered it difficult to drug to date, su ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
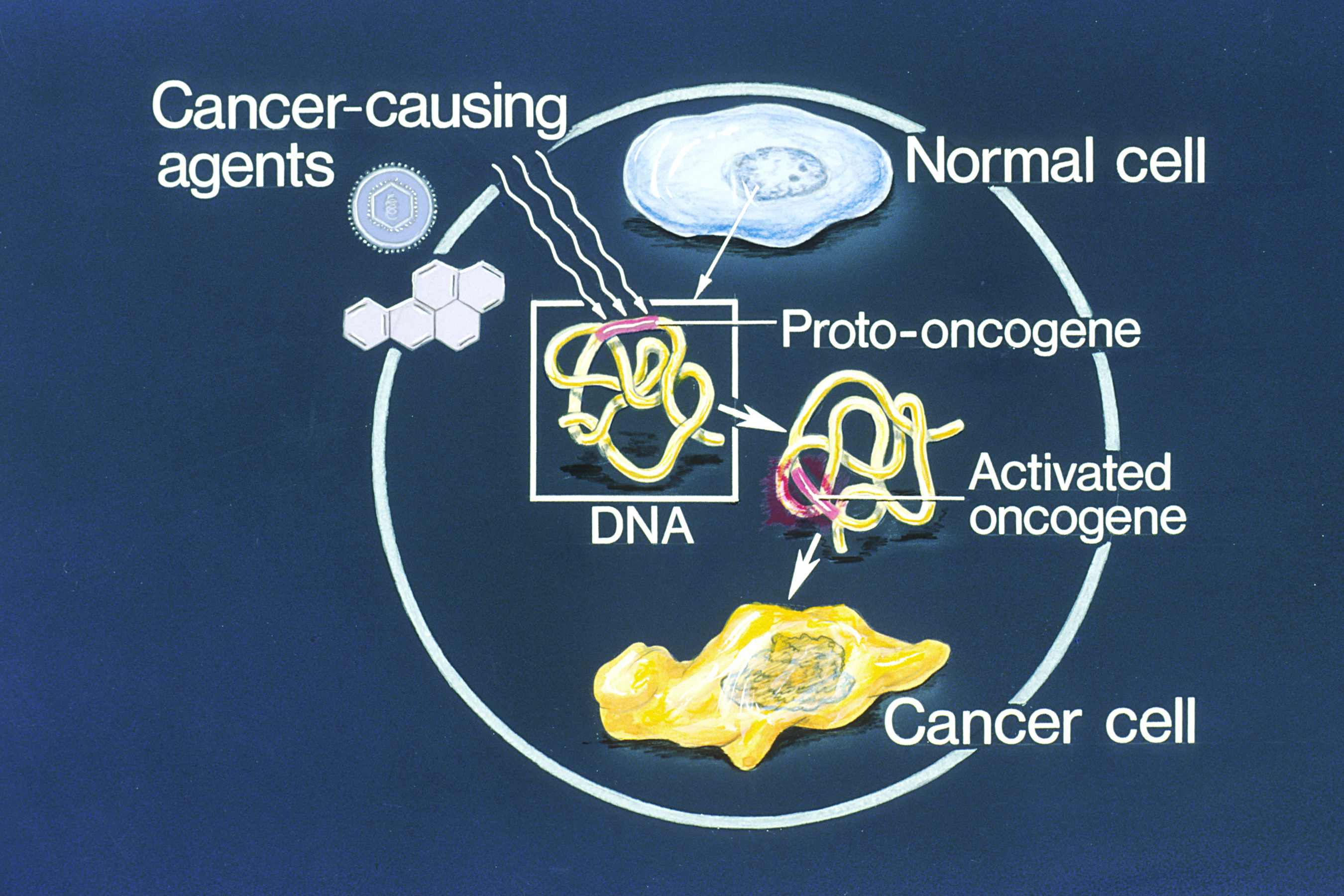
Oncogenes
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.Kimball's Biology Pages. "Oncogenes" Free full text Most normal cells undergo a preprogrammed rapid cell death () if critical functions are altered and then malfunction. Activated oncogenes can cause those cells designated for apoptosis to survive and proliferate instead. Most oncogenes began as proto-oncogenes: normal genes involved in cell growth and proliferation or inhibition of apoptosis. If, through mutation, normal genes promoting cellular growth are up-regulated (gain-of-function mutation), they predispose the cel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are approximately 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CAAT Box
In molecular biology, a CCAAT box (also sometimes abbreviated a CAAT box or CAT box) is a distinct pattern of nucleotides with GGCCAATCT consensus sequence that occur upstream by 60–100 bases to the initial transcription site. The CAAT box signals the binding site for the RNA transcription factor, and is typically accompanied by a conserved consensus sequence. It is an invariant DNA sequence at about minus 70 base pairs from the origin of transcription in many eukaryotic promoters. Genes that have this element seem to require it for the gene to be transcribed in sufficient quantities. It is frequently absent from genes that encode proteins used in virtually all cells. This box along with the GC box is known for binding general transcription factors. Both of these consensus sequences belong to the regulatory promoter. Full gene expression occurs when transcription activator proteins bind to each module within the regulatory promoter. Protein specific binding is required for t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TATA Box
In molecular biology, the TATA box (also called the Goldberg–Hogness box) is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The bacterial homolog of the TATA box is called the Pribnow box which has a shorter consensus sequence. The TATA box is considered a non-coding DNA sequence (also known as a cis-regulatory element). It was termed the "TATA box" as it contains a consensus sequence characterized by repeating T and A base pairs. How the term "box" originated is unclear. In the 1980s, while investigating nucleotide sequences in mouse genome loci, the Hogness box sequence was found and "boxed in" at the -31 position. When consensus nucleotides and alternative ones were compared, homologous regions were "boxed" by the researchers. The boxing in of sequences sheds light on the origin of the term "box". The TATA box was first identified in 1978 as a component of eukaryotic promoters. Transcription is initiated at the TATA box in TA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Introns
An intron is any Nucleic acid sequence, nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron [i.e., gene] ... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978) The term ''intron'' refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA Transcription (genetics), transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons. Introns are found in the genes of most eukaryotes and many eukaryotic viruses, and they can be located in both protein-coding genes and genes that function as RNA (Non-coding RNA, noncoding genes). There are f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |