|
Ribosylpyrimidine Nucleosidase
In enzymology, a ribosylpyrimidine nucleosidase () is an enzyme that catalyzes the chemical reaction :a pyrimidine nucleoside + H2O \rightleftharpoons D-ribose + a pyrimidine base Thus, the two substrates of this enzyme are pyrimidine nucleoside and H2O, whereas its two products are D-ribose and pyrimidine base. This enzyme belongs to the family of hydrolases, specifically those glycosylases that hydrolyse N-glycosyl compounds. The systematic name of this enzyme class is pyrimidine-nucleoside ribohydrolase. Other names in common use include N-ribosylpyrimidine nucleosidase, pyrimidine nucleosidase, N-ribosylpyrimidine ribohydrolase, pyrimidine nucleoside hydrolase, RihB, YeiK, and nucleoside ribohydrolase. This enzyme participates in purine metabolism and pyrimidine metabolism. Structural studies As of late 2007, two structures A structure is an arrangement and organization of interrelated elements in a material object or system, or the object or system so organized. Mate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymology
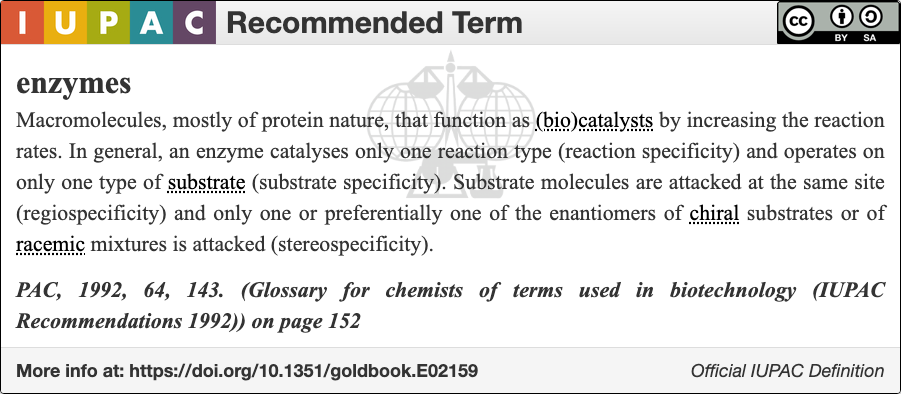
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrolase
In biochemistry, hydrolases constitute a class of enzymes that commonly function as biochemical catalysts that use water to break a chemical bond: :\ce \quad \xrightarrowtext\quad \ce This typically results in dividing a larger molecule into smaller molecules. Some common examples of hydrolase enzymes are esterases including lipases, phosphatases, glycosidases, peptidases, and nucleosidases. Esterases cleave ester bonds in lipids and phosphatases cleave phosphate groups off molecules. An example of crucial esterase is acetylcholine esterase, which assists in transforming the neuron impulse into the acetate group after the hydrolase breaks the acetylcholine into choline and acetic acid. Acetic acid is an important metabolite in the body and a critical intermediate for other reactions such as glycolysis. Lipases hydrolyze glycerides. Glycosidases cleave sugar molecules off carbohydrates and peptidases hydrolyze peptide bonds. Nucleosidases hydrolyze the bonds of nucleo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structure (journal)
''Structure'' is a monthly peer-reviewed scientific journal established in September 1993 by Wayne Hendrickson, Carl-Ivar Brändén, and Alan R. Fersht. It focuses on structural biology, studies of macromolecular structure, and related issues. In early 1999, the journal merged with ''Folding & Design'' and the name changed to ''Structure with Folding & Design''. In 2001, the journal reverted to ''Structure''. The journal is published by Cell Press and Christopher D. Lima and Andrej Sali served as editors-in-chief An editor-in-chief (EIC), also known as lead editor or chief editor, is a publication's editorial leader who has final responsibility for its operations and policies. The editor-in-chief heads all departments of the organization and is held accoun ... from 2003 to October 2021. The journal is now edited by an in-house team at Cell Press, with Karin Kühnel as editor-in-chhief. External links * Biochemistry journals Cell Press academic journals Academic journ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Acta Crystallogr
''Acta Crystallographica'' is a series of peer-reviewed scientific journals, with articles centred on crystallography, published by the International Union of Crystallography (IUCr). Originally established in 1948 as a single journal called ''Acta Crystallographica'', there are now six independent ''Acta Crystallographica'' titles: *'' Acta Crystallographica Section A: Foundations and Advances'' *'' Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials'' *'' Acta Crystallographica Section C: Structural Chemistry'' *'' Acta Crystallographica Section D: Structural Biology'' *'' Acta Crystallographica Section E: Crystallographic Communications'' *'' Acta Crystallographica Section F: Structural Biology Communications'' ''Acta Crystallographica'' has been noted for the high quality of the papers that it produces, as well as the large impact that its papers have had on the field of crystallography. The current six journals form part of the journal por ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). This structural data is obtained and deposited by biologists and biochemists worldwide through the use of experimental methodologies such as X-ray crystallography, Nuclear magnetic resonance spectroscopy of proteins, NMR spectroscopy, and, increasingly, cryo-electron microscopy. All submitted data are reviewed by expert Biocuration, biocurators and, once approved, are made freely available on the Internet under the CC0 Public Domain Dedication. Global access to the data is provided by the websites of the wwPDB member organizations (PDBe, PDBj, RCSB PDB, and BMRB). The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tertiary Structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the backbone may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure. A number of these structures may bind to each other, forming a quaternary structure. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypeptide chains and amino acid side chains, it was Dorothy Maud Wrinch who incorporated geometry into the prediction of protein structures. Wrinch demon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrimidine Metabolism
Pyrimidine biosynthesis occurs both in the body and through organic synthesis. ''De novo'' biosynthesis of pyrimidine ''De Novo'' biosynthesis of a pyrimidine is catalyzed by three gene products CAD, DHODH and UMPS. The first three enzymes of the process are all coded by the same gene in CAD which consists of carbamoyl phosphate synthetase II, aspartate carbamoyltransferase and dihydroorotase. Dihydroorotate dehydrogenase (DHODH) unlike CAD and UMPS is a mono-functional enzyme and is localized in the mitochondria. UMPS is a bifunctional enzyme consisting of orotate phosphoribosyltransferase (OPRT) and orotidine monophosphate decarboxylase (OMPDC). Both, CAD and UMPS are localized around the mitochondria, in the cytosol. In Fungi, a similar protein exists but lacks the dihydroorotase function: another protein catalyzes the second step. In other organisms (Bacteria, Archaea and the other Eukaryota), the first three steps are done by three different enzymes. Pyrimidine c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Purine Metabolism
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms. Biosynthesis Purines are biologically synthesized as nucleotides and in particular as ribotides, i.e. bases attached to ribose 5-phosphate. Both adenine and guanine are derived from the nucleotide inosine monophosphate (IMP), which is the first compound in the pathway to have a completely formed purine ring system. IMP Inosine monophosphate is synthesized on a pre-existing ribose-phosphate through a complex pathway (as shown in the figure on the right). The source of the carbon and nitrogen atoms of the purine ring, 5 and 4 respectively, come from multiple sources. The amino acid glycine contributes all its carbon (2) and nitrogen (1) atoms, with additional nitrogen atoms from glutamine (2) and aspartic acid (1), and additional carbon atoms from formyl groups (2), which are transferred from the coenzyme tetrahydrofolate as 10-formyltetrahydrofolate, and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
Enzymes are listed here by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system: :Oxidoreductases (EC 1) ( Oxidoreductase) * Dehydrogenase * Luciferase * DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) ** Homoserine dehydrogenase ** Aminopropanol oxidoreductase ** Diacetyl reductase ** Glycerol dehydrogenase ** Propanediol-phosphate dehydrogenase ** glycerol-3-phoshitiendopene dehydrogenase (NAD+) ** D-xylulose reductase ** L-xylulose reductase ** Lactate dehydrogenase ** Malate dehydrogenase ** Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) ** Glucose oxidase ** L-gulonolactone oxidase ** Thiamine oxidase ** Xanthine oxidase * EC 1.1.4 (with a disulfide as accep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrimidine Base
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The other diazines are pyrazine (nitrogen atoms at the 1 and 4 positions) and pyridazine (nitrogen atoms at the 1 and 2 positions). In nucleic acids, three types of nucleobases are pyrimidine derivatives: cytosine (C), thymine (T), and uracil (U). Occurrence and history The pyrimidine ring system has wide occurrence in nature as substituted and ring fused compounds and derivatives, including the nucleotides cytosine, thymine and uracil, thiamine (vitamin B1) and alloxan. It is also found in many synthetic compounds such as barbiturates and the HIV drug zidovudine. Although pyrimidine derivatives such as alloxan were known in the early 19th century, a laboratory synthesis of a pyrimidine was not carried out until 1879, when Grimaux reported the p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
D-ribose
Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally occurring form, , is a component of the ribonucleotides from which RNA is built, and so this compound is necessary for coding, decoding, regulation and expression of genes. It has a structural analog, deoxyribose, which is a similarly essential component of DNA. is an unnatural sugar that was first prepared by Emil Fischer and Oscar Piloty in 1891. It was not until 1909 that Phoebus Levene and Walter Jacobs recognised that was a natural product, the enantiomer of Fischer and Piloty's product, and an essential component of nucleic acids. Fischer chose the name "ribose" as it is a partial rearrangement of the name of another sugar, arabinose, of which ribose is an epimer at the 2' carbon; both names also relate to gum arabic, from which arabinose was first isolated and from which they prepared . Like most sugars, ribose exists as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |