|
Rare Biosphere
Rare biosphere refers to a large number of rare species of microbial life, i.e. bacteria, archaea and fungi, that can be found in very low concentrations in an environment. Microbial ecosystems Changes in the biodiversity of an ecosystem, whether marine or terrestrial, may affect its efficiency and function. Climate change or other anthropogenic perturbations can decrease productivity and disrupt global biogeochemical cycles. The possible ramifications of such changes are not well characterized or understood, and up to a point redundancy in an ecosystem may protect it from disruption. The dynamics of microbial ecosystems are tightly coupled to biogeochemical processes. For example, in the marine microbial loop, bacteria decompose organics and recycle nutrients such as nitrogen for other organisms such as phytoplankton to use. A reduction in recycled nitrogen would limit the production rate of phytoplankton, in turn limiting the growth of grazers, with effects throughout the f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit the air, soil, water, Hot spring, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria play a vital role in many stages of the nutrient cycle by recycling nutrients and the nitrogen fixation, fixation of nitrogen from the Earth's atmosphere, atmosphere. The nutrient cycle includes the decomposition of cadaver, dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, suc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
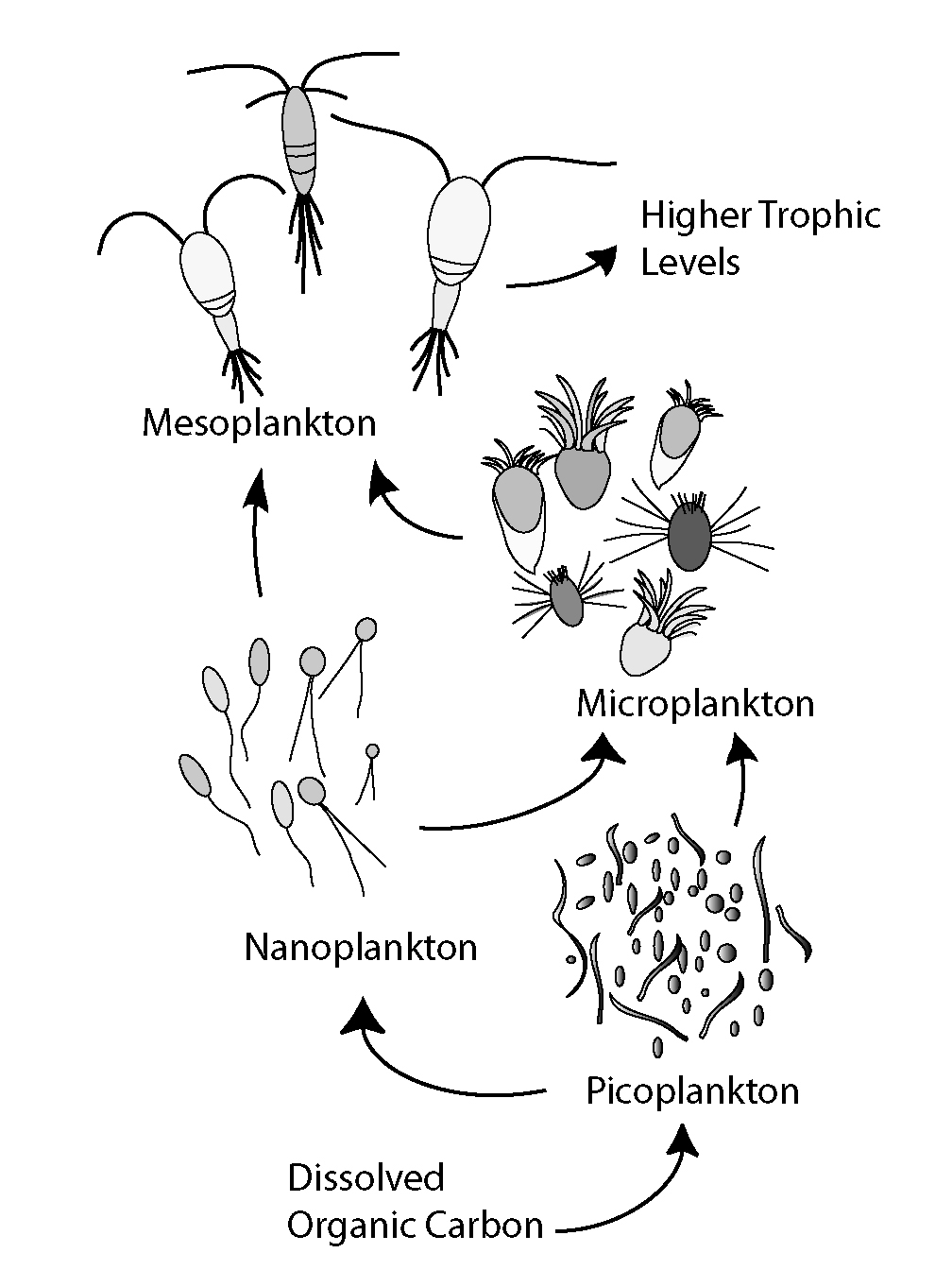
Phytoplankton
Phytoplankton () are the autotrophic (self-feeding) components of the plankton community and a key part of ocean and freshwater Aquatic ecosystem, ecosystems. The name comes from the Greek language, Greek words (), meaning 'plant', and (), meaning 'wanderer' or 'drifter'. Phytoplankton obtain their energy through photosynthesis, as trees and other plants do on land. This means phytoplankton must have light from the sun, so they live in the well-lit surface layers (euphotic zone) of oceans and lakes. In comparison with terrestrial plants, phytoplankton are distributed over a larger surface area, are exposed to less seasonal variation and have markedly faster turnover rates than trees (days versus decades). As a result, phytoplankton respond rapidly on a global scale to climate variations. Phytoplankton form the base of marine and freshwater food webs and are key players in the global carbon cycle. They account for about half of global photosynthetic activity and at least half o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Primer (molecular Biology)
A primer is a short, single-stranded nucleic acid used by all living organisms in the initiation of DNA synthesis. A synthetic primer is a type of oligo, short for oligonucleotide. DNA polymerases (responsible for DNA replication) are only capable of adding nucleotides to the 3’-end of an existing nucleic acid, requiring a primer be bound to the template before DNA polymerase can begin a complementary strand. DNA polymerase adds nucleotides after binding to the RNA primer and synthesizes the whole strand. Later, the RNA strands must be removed accurately and replaced with DNA nucleotides. This forms a gap region known as a nick that is filled in using a ligase. The removal process of the RNA primer requires several enzymes, such as Fen1, Lig1, and others that work in coordination with DNA polymerase, to ensure the removal of the RNA nucleotides and the addition of DNA nucleotides. Living organisms use solely RNA primers, while laboratory techniques in biochemistry and mole ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Shotgun Sequencing
In genetics, shotgun sequencing is a method used for sequencing random DNA strands. It is named by analogy with the rapidly expanding, quasi-random shot grouping of a shotgun. The Sanger sequencing#Method, chain-termination method of DNA sequencing ("Sanger sequencing") can only be used for short DNA strands of 100 to 1000 base pairs. Due to this size limit, longer sequences are subdivided into smaller fragments that can be sequenced separately, and these sequences are sequence assembly, assembled to give the overall sequence. In shotgun sequencing, DNA is broken up randomly into numerous small segments, which are sequenced using the chain termination method to obtain ''reads''. Multiple overlapping reads for the target DNA are obtained by performing several rounds of this fragmentation and sequencing. Computer programs then use the overlapping ends of different reads to assemble them into a continuous sequence. Shotgun sequencing was one of the precursor technologies that was resp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Operational Taxonomic Unit
An operational taxonomic unit (OTU) is an operational definition used to classify groups of closely related individuals. The term was originally introduced in 1963 by Robert R. Sokal and Peter H. A. Sneath in the context of numerical taxonomy, where an "operational taxonomic unit" is simply the group of organisms currently being studied. In this sense, an OTU is a pragmatic definition to group individuals by similarity, equivalent to but not necessarily in line with classical Linnaean taxonomy or modern evolutionary taxonomy. Nowadays, however, the term "OTU" is commonly used in a different context and refers to clusters of (uncultivated or unknown) organisms, grouped by DNA sequence similarity of a specific taxonomic marker gene (originally coined as mOTU; molecular OTU). In other words, OTUs are pragmatic proxies for "species" (microbial or metazoan) at different taxonomic levels, in the absence of traditional systems of biological classification as are available for macroscopic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RRNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form SSU rRNA, small and LSU rRNA, large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and Translation (biology), translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins, though this ratio differs between Prokaryote, prokaryotes and Eukaryote, eukaryotes. Structure Although the primary structure of rRNA sequences can vary across organisms, Base pair, base-pairing within these sequ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
High Throughput Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid advancements in DNA sequencing technology ha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Marine Biological Laboratory
The Marine Biological Laboratory (MBL) is an international center for research and education in biological and environmental science. Founded in Woods Hole, Massachusetts, in 1888, the MBL is a private, nonprofit institution that was independent for most of its history, but became officially affiliated with the University of Chicago on July 1, 2013. It also collaborates with numerous other institutions. As of 2024, 63 Nobel Prize winners have been affiliated with MBL as students, faculty members or researchers. In addition, since 1960, there have been 137 Howard Hughes Medical Institute investigators, early career scientists, international researchers, and professors; 319 members of the National Academy of Sciences; and 260 Members of the American Academy of Arts and Sciences who have been affiliated with the lab. History 19th century The Marine Biological Laboratory grew from the vision of several Bostonians and Spencer Fullerton Baird, the United States' first Fish Commiss ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitchell Sogin
Mitchell Sogin is an American microbiologist. He is a distinguished senior scientist at the Marine Biological Laboratory in Woods Hole, Massachusetts. His research investigates the evolution, diversity,and distribution of single-celled organisms. Early life and education Sogin grew up in Chicago and went to the University of Illinois on a swimming scholarship, intending to become a doctor. Instead he continued on at the university to complete a MS in Industrial Microbiology under Z. John Ordal in 1967 and a PhD in Microbiology and Molecular Biology under Carl R. Woese in 1972. Career Sogin obtained a BS in Chemistry and Microbiology at the University of Illinois, Urbana in 1967. He went on to join the staff at National Jewish Health, Denver, Colorado where he was an NIH Postdoctoral Fellow in the lab of Norman R. Pace from 1972-1976 and then a senior staff member from 1976-1989. Sogin became a professor at the University of Colorado Health Sciences Center in 1980 and in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, Genographic Project, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid advancements in DNA seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sanger Sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Frederick Sanger and colleagues in 1977, it became the most widely used sequencing method for approximately 40 years. An automated instrument using slab gel electrophoresis and fluorescent labels was first commercialized by Applied Biosystems in March 1987. Later, automated slab gels were replaced with automated capillary array electrophoresis. Recently, higher volume Sanger sequencing has been replaced by next generation sequencing methods, especially for large-scale, automated genome analyses. However, the Sanger method remains in wide use for smaller-scale projects and for validation of deep sequencing results. It still has the advantage over short-read sequencing technologies (like Illumina) in that it can produce DNA sequence reads of > ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |