|
Poly-A
Poly(A)-binding protein (PAB or PABP) is an RNA-binding protein which triggers the binding of eukaryotic initiation factor 4 complex (eIF4G) directly to the poly(A) tail of mRNA which is 200-250 nucleotides long. The poly(A) tail is located on the 3' end of mRNA and was discovered by Mary Edmonds, who also characterized the poly-A polymerase enzyme that generates the poly(a) tail. The binding protein is also involved in mRNA precursors by helping polyadenylate polymerase add the poly(A) nucleotide tail to the pre-mRNA before translation. The nuclear isoform selectively binds to around 50 nucleotides and stimulates the activity of polyadenylate polymerase by increasing its affinity towards RNA. Poly(A)-binding protein is also present during stages of mRNA metabolism including nonsense-mediated decay and nucleocytoplasmic trafficking. The poly(A)-binding protein may also protect the tail from degradation and regulate mRNA production. Without these two proteins in-tandem, then th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mary Edmonds
Mary P. Edmonds (May 7, 1922 – April 16, 2005) was an American biochemist who made key discoveries regarding the processing of messenger RNA (mRNA). She spent most of her career at the University of Pittsburgh. Education and career Edmonds was born May 7, 1922, in Racine, Wisconsin. She received a bachelor's degree from Milwaukee-Downer College in 1943, a master's degree from Wellesley College in 1945, and a Ph.D. from the University of Pennsylvania in 1951. Following her Ph.D., she was a postdoctoral researcher at University of Illinois (1950-1952) and University of Wisconsin (1952-1955), and then joined Montefiore Hospital in Pittsburg as a research associate from 1955 until 1965. Starting in 1962 she held appointments at the University of Pittsburgh, initially as adjunct and research professor positions, until she joined the faculty in 1971 and was promoted to professor in 1976. Edmonds took emeritus status in 1992, and died of complications related to a heart attack on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Poly(A)-binding Protein
Poly(A)-binding protein (PAB or PABP) is an RNA-binding protein which triggers the binding of eukaryotic initiation factor 4 complex (eIF4G) directly to the poly(A) tail of mRNA which is 200-250 nucleotides long. The poly(A) tail is located on the 3' end of mRNA and was discovered by Mary Edmonds, who also characterized the poly-A polymerase enzyme that generates the poly(a) tail. The binding protein is also involved in mRNA precursors by helping polyadenylate polymerase add the poly(A) nucleotide tail to the pre-mRNA before translation. The nuclear isoform selectively binds to around 50 nucleotides and stimulates the activity of polyadenylate polymerase by increasing its affinity towards RNA. Poly(A)-binding protein is also present during stages of mRNA metabolism including nonsense-mediated decay and nucleocytoplasmic trafficking. The poly(A)-binding protein may also protect the tail from degradation and regulate mRNA production. Without these two proteins in-tandem, then the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Poly(A) Tail
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression. The process of polyadenylation begins as the transcription of a gene terminates. The 3′-most segment of the newly made pre-mRNA is first cleaved off by a set of proteins; these proteins then synthesize the poly(A) tail at the RNA's 3′ end. In some genes these proteins add a poly(A) tail at one of several possible sites. Therefore, polyadenylation can produce more than one transcript from a single gene (alternative polyadenylation), similar to alternative splicing. The poly(A) tail is important for the nucl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotic Translation
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: gene translation, elongation, termination, and recapping. Initiation Translation initiation is the process by which the ribosome and its associated factors bind to an mRNA and are assembled at the start codon. This process is defined as either cap-dependent, in which the ribosome binds initially at the 5' cap and then travels to the stop codon, or as cap-independent, where the ribosome does not initially bind the 5' cap. Cap-dependent initiation Initiation of translation usually involves the interaction of certain key proteins, the initiation factors, with a special tag bound to the 5'-end of an mRNA molecule, the 5' cap, as well as with the 5' UTR. These proteins bind the small (40S) ribosomal subunit and hold the mRNA in place. eIF3 is associated with the 40S ribosomal subunit and plays a role in keeping the large (60S) ribosomal s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Untranslated Region
In molecular genetics, an untranslated region (or UTR) refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the 5' side, it is called the 5' UTR (or leader sequence), or if it is found on the 3' side, it is called the 3' UTR (or trailer sequence). mRNA is RNA that carries information from DNA to the ribosome, the site of protein synthesis (translation) within a cell. The mRNA is initially transcribed from the corresponding DNA sequence and then translated into protein. However, several regions of the mRNA are usually not translated into protein, including the 5' and 3' UTRs. Although they are called untranslated regions, and do not form the protein-coding region of the gene, uORFs located within the 5' UTR can be translated into peptides. The 5' UTR is upstream from the coding sequence. Within the 5' UTR is a sequence that is recognized by the ribosome which allows the ribosome to bind and initiate translatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conserved Sequence
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domain (biology), domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA#History, DNA in heredity, and observations by Frederick Sanger of variation betwee ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
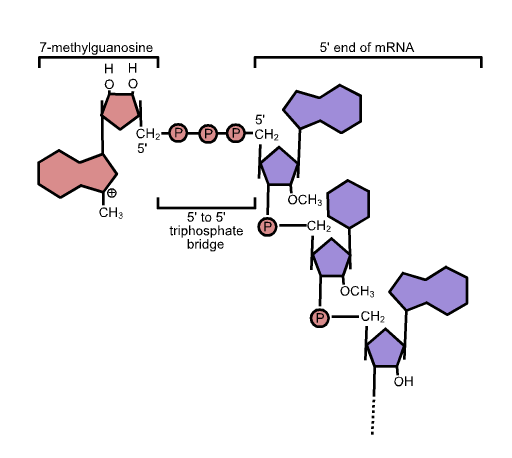
5' Cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation of stable and mature messenger RNA able to undergo translation during protein synthesis. Mitochondrial mRNA and chloroplastic mRNA are not capped. Structure In eukaryotes, the 5′ cap (cap-0), found on the 5′ end of an mRNA molecule, consists of a guanine nucleotide connected to mRNA via an unusual 5′ to 5′ triphosphate linkage. This guanosine is methylated on the 7 position directly after capping ''in vivo'' by a methyltransferase. It is referred to as a 7-methylguanylate cap, abbreviated m7G. In multicellular eukaryotes and some viruses, further modifications exist, including the methylation of the 2′ hydroxy-groups of the first 2 ribose sugars of the 5′ end of the mRNA. cap-1 has a methylated 2′-hydroxy group ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EIF4E
Eukaryotic translation initiation factor 4E, also known as eIF4E, is a protein that in humans is encoded by the ''EIF4E'' gene. Structure and function Most eukaryotic cellular mRNAs are blocked at their 5'-ends with the 7-methyl- guanosine five-prime cap structure, m7GpppX (where X is any nucleotide). This structure is involved in several cellular processes including enhanced translational efficiency, splicing, mRNA stability, and RNA nuclear export. eIF4E is a eukaryotic translation initiation factor involved in directing ribosomes to the cap structure of mRNAs. It is a 24-kD polypeptide that exists as both a free form and as part of the eIF4F pre-initiation complex. Almost all cellular mRNA require eIF4E in order to be translated into protein. The eIF4E polypeptide is the rate-limiting component of the eukaryotic translation apparatus and is involved in the mRNA-ribosome binding step of eukaryotic protein synthesis. The other subunits of eIF4F are a 47-kD polypeptide, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EIF4F
Eukaryotic initiation factor 4F (eIF4F) is a heterotrimeric protein complex that binds the 5' cap of messenger RNAs (mRNAs) to promote eukaryotic translation initiation. The eIF4F complex is composed of three non-identical subunits: the DEAD-box RNA helicase eIF4A, the cap-binding protein eIF4E, and the large "scaffold" protein eIF4G. The mammalian eIF4F complex was first described in 1983, and has been a major area of study into the molecular mechanisms of cap-dependent translation initiation ever since. Function eIF4F is important for recruiting the small ribosomal subunit (40S) to the 5' cap of mRNAs during cap-dependent translation initiation. Components of the complex are also involved in cap-independent translation initiation; for instance, certain viral proteases cleave eIF4G to remove the eIF4E-binding region, thus inhibiting cap-dependent translation. Structure Structures of eIF4F components have been solved individually and as partial complexes by a variety ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EIF4G
Eukaryotic translation initiation factor 4 G (eIF4G) is a protein involved in eukaryotic translation initiation and is a component of the eIF4F cap-binding complex. Orthologs of eIF4G have been studied in multiple species, including humans, yeast, and wheat. However, eIF4G is exclusively found in domain Eukarya, and not in domains Bacteria or Archaea, which do not have capped mRNA. As such, eIF4G structure and function may vary between species, although the human EIF4G1 has been the focus of extensive studies. (Other human paralogs are EIF4G2 and EIF4G3.) Across species, eIF4G strongly associates with eIF4E, the protein that directly binds the mRNA cap. Together with the RNA helicase protein eIF4A, these form the eIF4F complex. Within the cell eIF4G is found primarily in the cytoplasm, usually bound to eIF4E; however, it is also found in the nucleus, where its function is unknown. It may have a role in nonsense-mediated decay. History eIF4G stands for eukaryotic initia ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytosol
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells ( intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into many compartments. In the eukaryotic cell, the cytosol is surrounded by the cell membrane and is part of the cytoplasm, which also comprises the mitochondria, plastids, and other organelles (but not their internal fluids and structures); the cell nucleus is separate. The cytosol is thus a liquid matrix around the organelles. In prokaryotes, most of the chemical reactions of metabolism take place in the cytosol, while a few take place in membranes or in the periplasmic space. In eukaryotes, while many metabolic pathways still occur in the cytosol, others take place within organelles. The cytosol is a complex mixture of substances dissolved in water. Although water forms the large majority of the cytosol, its structure and prop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carboxylic Acid
In organic chemistry, a carboxylic acid is an organic acid that contains a carboxyl group () attached to an R-group. The general formula of a carboxylic acid is or , with R referring to the alkyl, alkenyl, aryl, or other group. Carboxylic acids occur widely. Important examples include the amino acids and fatty acids. Deprotonation of a carboxylic acid gives a carboxylate anion. Examples and nomenclature Carboxylic acids are commonly identified by their trivial names. They at oftentimes have the suffix ''-ic acid''. IUPAC-recommended names also exist; in this system, carboxylic acids have an ''-oic acid'' suffix. For example, butyric acid (C3H7CO2H) is butanoic acid by IUPAC guidelines. For nomenclature of complex molecules containing a carboxylic acid, the carboxyl can be considered position one of the parent chain even if there are other substituents, such as 3-chloropropanoic acid. Alternately, it can be named as a "carboxy" or "carboxylic acid" substituent on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
-binding_protein.png)