|
Phylogenomics
Phylogenomics is the intersection of the fields of evolution and genomics. The term has been used in multiple ways to refer to analysis that involves genome data and evolutionary reconstructions. It is a group of techniques within the larger fields of phylogenetics and genomics. Phylogenomics draws information by comparing entire genomes, or at least large portions of genomes. Phylogenetics compares and analyzes the sequences of single genes, or a small number of genes, as well as many other types of data. Four major areas fall under phylogenomics: * Prediction of gene function * Establishment and clarification of evolutionary relationships * Gene family evolution * Prediction and retracing lateral gene transfer. The ultimate goal of phylogenomics is to reconstruct the evolutionary history of species through their genomes. This history is usually inferred from a series of genomes by using a genome evolution model and standard statistical inference methods (e.g. Bayesian inference or ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Evolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, resulting in certain characteristics becoming more or less common within a population over successive generations. The process of evolution has given rise to biodiversity at every level of biological organisation. The scientific theory of evolution by natural selection was conceived independently by two British naturalists, Charles Darwin and Alfred Russel Wallace, in the mid-19th century as an explanation for why organisms are adapted to their physical and biological environments. The theory was first set out in detail in Darwin's book ''On the Origin of Species''. Evolution by natural selection is established by observable facts about living organisms: (1) more offspring are often produced than can possibly survive; (2) phenotypic variatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Species
A species () is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. It is the basic unit of Taxonomy (biology), classification and a taxonomic rank of an organism, as well as a unit of biodiversity. Other ways of defining species include their karyotype, DNA sequence, morphology (biology), morphology, behaviour, or ecological niche. In addition, palaeontologists use the concept of the chronospecies since fossil reproduction cannot be examined. The most recent rigorous estimate for the total number of species of eukaryotes is between 8 and 8.7 million. About 14% of these had been described by 2011. All species (except viruses) are given a binomial nomenclature, two-part name, a "binomen". The first part of a binomen is the name of a genus to which the species belongs. The second part is called the specific name (zoology), specific name or the specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Supertree
A supertree is a single phylogenetic tree assembled from a combination of smaller phylogenetic trees, which may have been assembled using different datasets (e.g. morphological and molecular) or a different selection of taxa. Supertree algorithms can highlight areas where additional data would most usefully resolve any ambiguities. The input trees of a supertree should behave as samples from the larger tree. Construction methods The construction of a supertree scales exponentially with the number of taxa included; therefore for a tree of any reasonable size, it is not possible to examine every possible supertree and weigh its success at combining the input information. Heuristic methods are thus essential, although these methods may be unreliable; the result extracted is often biased or affected by irrelevant characteristics of the input data. The most well known method for supertree construction is Matrix Representation with Parsimony (MRP), in which the input source trees ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural biology, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix (mathematics), matrix. Gaps are inserted between the Residue (chemistry), residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences such as calculating the Edit distance, distance cost between strings in a natural language, or to display financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence ali ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetics
In biology, phylogenetics () is the study of the evolutionary history of life using observable characteristics of organisms (or genes), which is known as phylogenetic inference. It infers the relationship among organisms based on empirical data and observed heritable traits of DNA sequences, protein amino acid sequences, and morphology. The results are a phylogenetic tree—a diagram depicting the hypothetical relationships among the organisms, reflecting their inferred evolutionary history. The tips of a phylogenetic tree represent the observed entities, which can be living taxa or fossils. A phylogenetic diagram can be rooted or unrooted. A rooted tree diagram indicates the hypothetical common ancestor of the taxa represented on the tree. An unrooted tree diagram (a network) makes no assumption about directionality of character state transformation, and does not show the origin or "root" of the taxa in question. In addition to their use for inferring phylogenetic pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microbial Phylogenetics
Microbial phylogenetics is the study of the manner in which various groups of microorganisms are genetically related. This helps to trace their evolution. To study these relationships biologists rely on comparative genomics, as physiology and comparative anatomy are not possible methods. History 1960s–1970s Microbial phylogenetics emerged as a field of study in the 1960s, scientists started to create genealogical trees based on differences in the order of amino acids of proteins and nucleotides of genes instead of using comparative anatomy and physiology. One of the most important figures in the early stage of this field is Carl Woese, who in his researches, focused on Bacteria, looking at RNA instead of proteins. More specifically, he decided to compare the small subunit ribosomal RNA (16rRNA) oligonucleotides. Matching oligonucleotides in different bacteria could be compared to one another to determine how closely the organisms were related. In 1977, after collecting and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Archaeopteryx (evolutionary Tree Visualization And Analysis)
Archaeopteryx is an interactive computer software program, written in Java, for viewing, editing, and analyzing phylogenetic trees. This type of program can be used for a variety of analyses of molecular data sets, but is particularly designed for phylogenomics Phylogenomics is the intersection of the fields of evolution and genomics. The term has been used in multiple ways to refer to analysis that involves genome data and evolutionary reconstructions. It is a group of techniques within the larger fields .... Besides tree description formats with limited expressiveness (such as Newick/New Hamphshire, Nexus), it also implements the phyloXML format. Archaeopteryx is the successor to Java program ''A Tree Viewer'' (ATV). References External linksphyloXML * ttp://www.phylosoft.org/archaeopteryx/ Archaeopteryx {{genomics-footer Phylogenetics software ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PhylomeDB
PhylomeDB is a public biological database for complete catalogs of gene phylogenies ( phylomes). It allows users to interactively explore the evolutionary history of genes through the visualization of phylogenetic trees and multiple sequence alignments. Moreover, phylomeDB provides genome-wide orthology and paralogy predictions which are based on the analysis of the phylogenetic trees. The automated pipeline used to reconstruct trees aims at providing a high-quality phylogenetic analysis of different genomes, including Maximum Likelihood tree inference, alignment trimming and evolutionary model testing. PhylomeDB includes also a public download section with the complete set of trees, alignments and orthology predictions, as well as a web API that facilitates cross linking trees from external sources. Finally, phylomeDB provides an advanced tree visualization interface based on the ETE toolkit,{{cite journal , last=Huerta-Cepas, J, author2= Dopazo, J, author3=Gabaldón, T , date=Jan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Taxonomy Database
The Genome Taxonomy Database (GTDB) is an online database that maintains information on a proposed nomenclature of prokaryotes, following a phylogenomic approach based on a set of conserved single-copy proteins. In addition to resolving paraphyletic groups, this method also reassigns taxonomic ranks algorithmically, updating names in both cases. Information for archaea was added in 2020, along with a species classification based on average nucleotide identity. Each update incorporates new genomes as well as automated and manual curation of the taxonomy. An open-source tool called GTDB-Tk is available to classify draft genomes into the GTDB hierarchy. The GTDB system, via GTDB-Tk, has been used to catalogue not-yet-named bacteria in the human gut microbiome and other metagenomic sources. The GTDB is incorporated into the '' Bergey's Manual of Systematics of Archaea and Bacteria'' in 2019 as its phylogenomic resource. Methodology The genomes used to construct the phyloge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plants+HC+SAR Megagroup
Diaphoretickes is a major group of eukaryotic organisms spanning over 400,000 species. The majority of the earth's biomass that carries out photosynthesis belongs to Diaphoretickes. In older classification systems, members of the Diaphoretickes were variously placed in the kingdoms Protozoa or Protista. Etymology The name Diaphoretickes derives (''diaforetikés'') meaning diverse, dissimilar, referring to the wide morphological and cellular diversity among members of this clade. History Eukaryotes, organisms whose cells contain a nucleus, have been traditionally grouped into four kingdoms: animals, plants, fungi and protists. In the late 20th century, molecular phylogenetic analyses revealed that protists are a paraphyletic assortment of many independent evolutionary lineages or clades, from which animals, fungi and plants evolved. However, the relationships between these clades remained difficult to assess due to technological limitations. Starting in the early 2000s, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cryptomonads
The cryptomonads (or cryptophytes) are a superclass of algae, most of which have plastids. They are traditionally considered a division of algae among phycologists, under the name of Cryptophyta. They are common in freshwater, and also occur in marine and brackish habitats. Each cell is around 10–50 μm in size and flattened in shape, with an anterior groove or pocket. At the edge of the pocket there are typically two slightly unequal flagella. Some may exhibit mixotrophy. They are classified as superclass Cryptomonada, which is divided into two classes: heterotrophic Goniomonadea and phototrophic Cryptophyceae. The two groups are united under three shared morphological characteristics: presence of a periplast, ejectisomes with secondary scroll, and mitochondrial cristae with flat tubules. Genetic studies as early as 1994 also supported the hypothesis that ''Goniomonas'' was sister to Cryptophyceae. A study in 2018 found strong evidence that the common ancestor of Cryp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
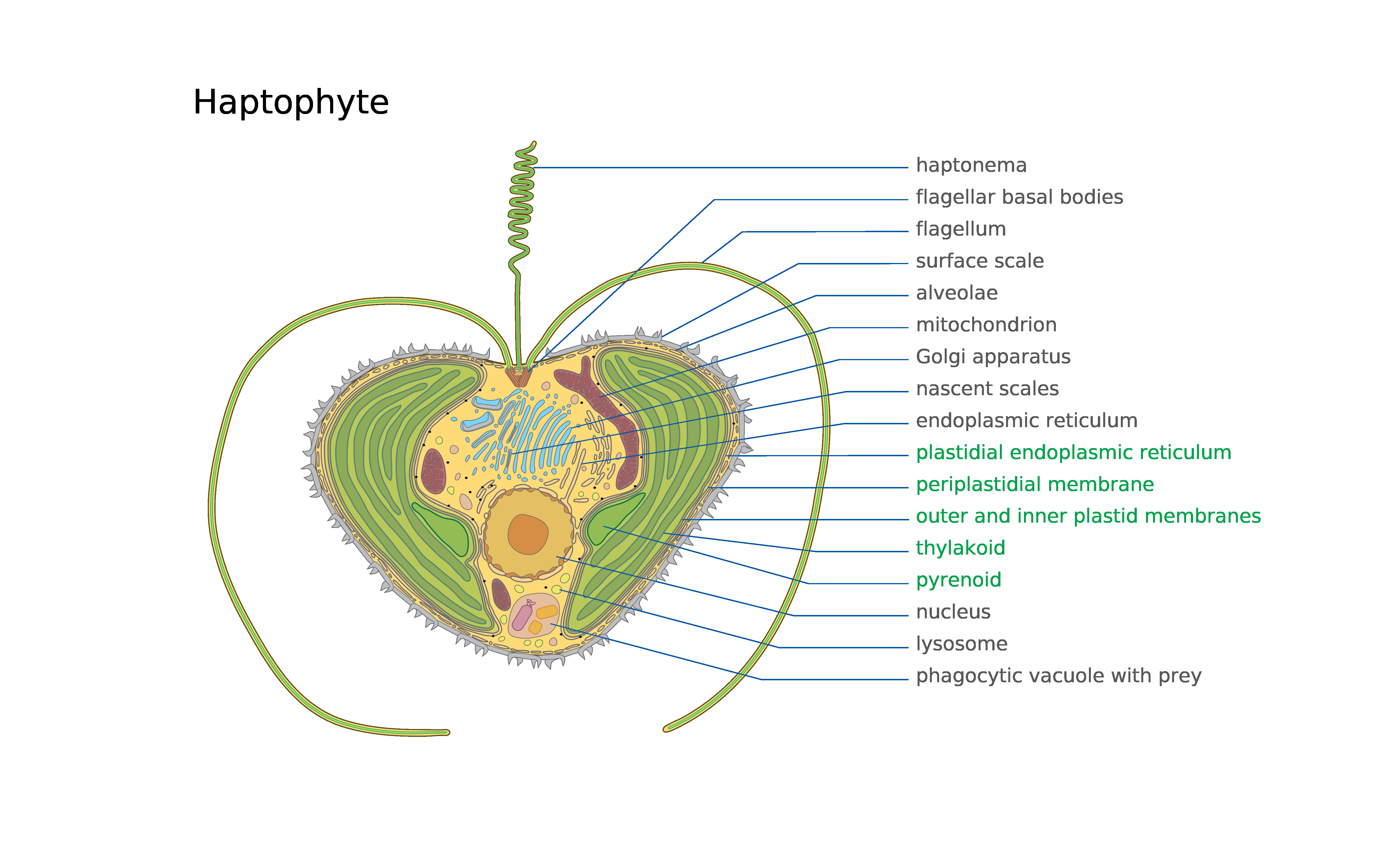
Haptophytes
The haptophytes, classified either as the Haptophyta, Haptophytina or Prymnesiophyta (named for ''Prymnesium''), are a clade of algae. The names Haptophyceae or Prymnesiophyceae are sometimes used instead. This ending implies classification at the class rank rather than as a division. Although the phylogenetics of this group has become much better understood in recent years, there remains some dispute over which rank is most appropriate. Characteristics The chloroplasts are pigmented similarly to those of the heterokonts, but the structure of the rest of the cell is different, so it may be that they are a separate line whose chloroplasts are derived from similar red algal endosymbionts. Haptophyte chloroplasts contain chlorophylls a, c1, and c2 but lack chlorophyll b. For carotenoids, they have beta-, alpha-, and gamma- carotenes. Like diatoms and brown algae, they have also fucoxanthin, an oxidized isoprenoid derivative that is likely the most important driver of their ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |