|
Phosphatidylinositol 3-kinase-related Kinase
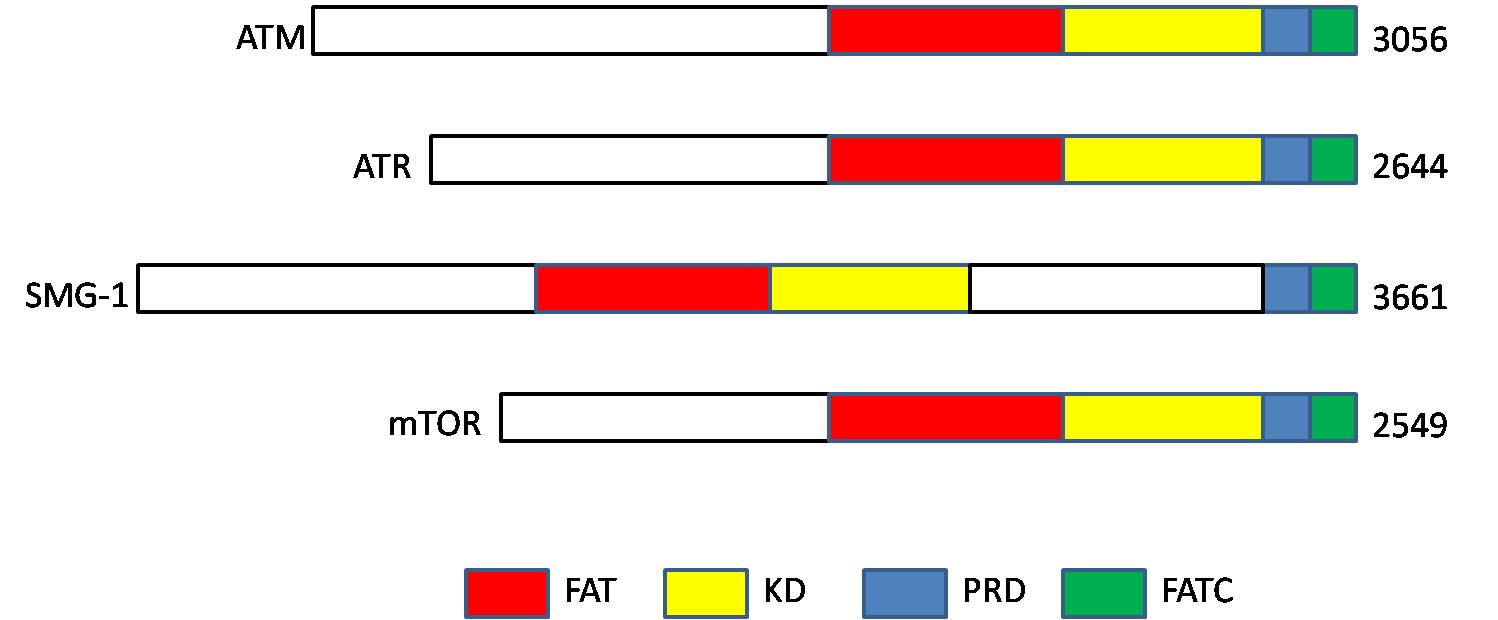
Phosphatidylinositol 3-kinase-related kinases (PIKKs) are a family of Ser/Thr-protein kinases with sequence similarity to phosphatidylinositol-3 kinases ( PI3Ks). Members The human PIKK family includes six members: Structure PIKKs proteins contain the following four domains: # N-terminus FRAP-ATM- TRRAP (FAT) domain, # kinase domain (KD; PI3_PI4_kinase), # PIKK- regulatory domain (PRD), and # C-terminus The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, carboxy tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein Proteins are large biomolecules and macromolecules that comp ... FAT-C-terminal (FATC) domain References External links Kinase Family PIKKaWikiKinome EC 2.7.11 Protein families {{protein-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine/threonine-specific Protein Kinase
A serine/threonine protein kinase () is a kinase enzyme, in particular a protein kinase, that phosphorylation, phosphorylates the hydroxyl, OH group of the amino acid, amino-acid residues serine or threonine, which have similar side chains. At least 350 of the 500+ human protein kinases are serine/threonine kinases (STK). In enzymology, the term ''serine/threonine protein kinase'' describes a class of enzymes in the family of transferases, that transfer phosphates to the oxygen atom of a serine or threonine side chain in proteins. This process is called phosphorylation. Protein phosphorylation in particular plays a significant role in a wide range of cellular processes and is a very important post-translational modification. The chemical reaction performed by these enzymes can be written as :ATP + a protein \rightleftharpoons ADP + a phosphoprotein Thus, the two substrate (biochemistry), substrates of this enzyme are adenosine triphosphate, ATP and a protein, whereas its ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphoinositide 3-kinase
Phosphoinositide 3-kinases (PI3Ks), also called phosphatidylinositol 3-kinases, are a family of enzymes involved in cellular functions such as cell growth, proliferation, differentiation, motility, survival and intracellular trafficking, which in turn are involved in cancer. PI3Ks are a family of related intracellular signal transducer enzymes capable of phosphorylating the 3 position hydroxyl group of the inositol ring of phosphatidylinositol (PtdIns). The pathway, with oncogene PIK3CA and tumor suppressor gene PTEN, is implicated in the sensitivity of cancer tumors to insulin and IGF1, and in calorie restriction. Discovery The discovery of PI3Ks by Lewis Cantley and colleagues began with their identification of a previously unknown phosphoinositide kinase associated with the polyoma middle T protein. They observed unique substrate specificity and chromatographic properties of the products of the lipid kinase, leading to the discovery that this phosphoinositide kinas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ataxia Telangiectasia Mutated
ATM serine/threonine kinase or Ataxia-telangiectasia mutated, symbol ATM, is a serine/threonine protein kinase that is recruited and activated by DNA repair#Double-strand breaks, DNA double-strand breaks (Canonical pathway, canonical pathway), oxidative stress, topoisomerase cleavage complexes, splicing intermediates, R-loops and in some cases by DNA repair, single-strand DNA breaks. It phosphorylates several key proteins that initiate activation of the DNA damage cell cycle checkpoint, checkpoint, leading to cell cycle arrest, DNA repair or apoptosis. Several of these targets, including p53, CHK2, BRCA1, NBS1 and H2AX are tumor suppressors. In 1995, the gene was discovered by Yosef Shiloh who named its product ATM since he found that its mutations are responsible for the disorder ataxia–telangiectasia#Cause, ataxia–telangiectasia. In 1998, the Shiloh and Michael B. Kastan, Kastan laboratories independently showed that ATM is a protein kinase whose activity is enhanced by DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ataxia Telangiectasia And Rad3 Related
Serine/threonine-protein kinase ATR, also known as ataxia telangiectasia and Rad3-related protein (ATR) or FRAP-related protein 1 (FRP1), is an enzyme that, in humans, is encoded by the ''ATR'' gene. It is a large kinase of about 301.66 kDa. ATR belongs to the phosphatidylinositol 3-kinase-related kinase protein family. ATR is activated in response to single strand breaks, and works with ATM to ensure genome integrity. Function ATR is a serine/threonine-specific protein kinase that is involved in sensing DNA damage and activating the DNA damage checkpoint, leading to cell cycle arrest in eukaryotes. ATR is activated in response to persistent single-stranded DNA, which is a common intermediate formed during DNA damage detection and repair. Single-stranded DNA occurs at stalled replication forks and as an intermediate in DNA repair pathways such as nucleotide excision repair and homologous recombination repair. ATR is activated during more persistent issues with DNA damage ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA-PKcs
DNA-dependent protein kinase catalytic subunit, also known as DNA-PKcs, is an enzyme that plays a crucial role in repairing DNA double-strand breaks and has a number of other DNA housekeeping functions. In humans it is encoded by the gene designated as ''PRKDC'' or ''XRCC7''. DNA-PKcs belongs to the phosphatidylinositol 3-kinase-related kinase protein family. The DNA-Pkcs protein is a serine/threonine protein kinase consisting of a single polypeptide chain of 4,128 amino acids. Function DNA-PKcs is the catalytic subunit of a nuclear DNA-dependent serine/threonine protein kinase called DNA-PK. The second component is the autoimmune antigen Ku. On its own, DNA-PKcs is inactive and relies on Ku to direct it to DNA ends and trigger its kinase activity. DNA-PKcs is required for the non-homologous end joining (NHEJ) pathway of DNA repair, which rejoins double-strand breaks. It is also required for V(D)J recombination, a process that utilizes NHEJ to promote immune system diversit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mammalian Target Of Rapamycin
The mammalian target of sirolimus, rapamycin (mTOR), also referred to as the mechanistic target of rapamycin, and sometimes called FK506-binding protein 12-rapamycin-associated protein 1 (FRAP1), is a kinase that in humans is encoded by the ''MTOR'' gene. mTOR is a member of the phosphatidylinositol 3-kinase-related kinase family of protein kinases. mTOR links with other proteins and serves as a core component of two distinct protein complexes, mTORC1, mTOR complex 1 and mTORC2, mTOR complex 2, which regulate different cellular processes. In particular, as a core component of both complexes, mTOR functions as a serine/threonine protein kinase that regulates cell growth, cell proliferation, cell motility, cell survival, protein synthesis, autophagy, and Transcription (genetics), transcription. As a core component of mTORC2, mTOR also functions as a tyrosine protein kinase that promotes the activation of insulin receptors and insulin-like growth factor 1 receptors. mTORC2 has also ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SMG1 (gene)
Serine/threonine-protein kinase SMG1 is an enzyme that in humans is encoded by the ''SMG1'' gene. SMG1 belongs to the phosphatidylinositol 3-kinase-related kinase protein family. Function This gene encodes a protein involved in nonsense-mediated mRNA decay (NMD) as part of the mRNA surveillance complex. The protein has kinase activity and is thought to function in NMD by phosphorylating the regulator of nonsense transcripts 1 protein. Alternative spliced transcript variants have been described, but their full-length natures have not been determined. Interactions SMG1 (gene) has been shown to interact with PRKCI and UPF1 Regulator of nonsense transcripts 1 (or Up-frameshift suppressor 1 homolog) is a protein that in humans is encoded by the ''UPF1'' gene. Function This gene encodes a protein that is part of a post-splicing multiprotein complex, the exon junct .... References Further reading * * * * * * * * * * * * * * EC 2.7.11 Genes on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and the ribosome creates the protein utilizing amino acids carried by transfer RNA (tRNA). This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, genetic inf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transformation/transcription Domain-associated Protein
Transformation/transcription domain-associated protein, also known as TRRAP, is a protein that in humans is encoded by the ''TRRAP'' gene. TRRAP belongs to the phosphatidylinositol 3-kinase-related kinase protein family. Function TRRAP is an adaptor protein, which is found in various multiprotein chromatin complexes with histone acetyltransferase activity (HAT), which in turn is responsible for epigenetic transcription activation. TRRAP has a central role in MYC (c-Myc) transcription activation, and also participates in cell transformation by MYC. It is required for p53/TP53-, E2F1-, and E2F4-mediated transcription activation. It is also involved in transcription activation mediated by the adenovirus E1A, a viral oncoprotein that deregulates transcription of key genes. TRRAP is also required for the mitotic checkpoint and normal cell cycle progression. The MRN complex (composed of MRE11, RAD50, and NBS1) is involved in the detection and repair of DNA double-strand breaks ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coactivator (genetics)
A coactivator is a type of transcriptional coregulator that binds to an activator (a transcription factor) to increase the rate of transcription of a gene or set of genes. The activator contains a DNA binding domain that binds either to a DNA promoter site or a specific DNA regulatory sequence called an enhancer. Binding of the activator-coactivator complex increases the speed of transcription by recruiting general transcription machinery to the promoter, therefore increasing gene expression. The use of activators and coactivators allows for highly specific expression of certain genes depending on cell type and developmental stage. Some coactivators also have histone acetyltransferase (HAT) activity. HATs form large multiprotein complexes that weaken the association of histones to DNA by acetylating the N-terminal histone tail. This provides more space for the transcription machinery to bind to the promoter, therefore increasing gene expression. Activators are found in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's Peptide, polypeptide chain that is self-stabilizing and that Protein folding, folds independently from the rest. Each domain forms a compact folded Protein tertiary structure, three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or Disulfide bond, disulfide bridges. Domains often form functional units, such as the calcium-binding EF-hand, EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimera (protein), chimeric ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |