|
P-bodies
In cellular biology, P-bodies, or processing bodies, are distinct foci formed by phase separation within the cytoplasm of a eukaryotic cell consisting of many enzymes involved in mRNA turnover. P-bodies are highly conserved structures and have been observed in somatic cells originating from vertebrates and invertebrates, plants and yeast. To date, P-bodies have been demonstrated to play fundamental roles in general mRNA decay, nonsense-mediated mRNA decay, adenylate-uridylate-rich element mediated mRNA decay, and microRNA (miRNA) induced mRNA silencing. Not all mRNAs which enter P-bodies are degraded, as it has been demonstrated that some mRNAs can exit P-bodies and re-initiate translation. Purification and sequencing of the mRNA from purified processing bodies showed that these mRNAs are largely translationally repressed upstream of translation initiation and are protected from 5' mRNA decay. P-bodies were originally proposed to be the sites of mRNA degradation in the cel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stress Granule
In cellular biology, stress granules are biomolecular condensates in the cytosol composed of proteins and RNA that assemble into 0.1–2 μm membraneless organelles when the cell is under stress. The mRNA molecules found in stress granules are stalled translation pre-initiation complexes associated with 40S ribosomal subunits, translation initiation factors, poly(A)+ mRNA and RNA-binding proteins (RBPs). While they are membraneless organelles, stress granules have been proposed to be associated with the endoplasmatic reticulum. There are also nuclear stress granules. This article is about the cytosolic variety. Proposed functions The function of stress granules remains largely unknown. Stress granules have long been proposed to have a function to protect RNA from harmful conditions, thus their appearance under stress. The accumulation of RNA into dense globules could keep them from reacting with harmful chemicals and safeguard the information coded in their RNA sequence ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
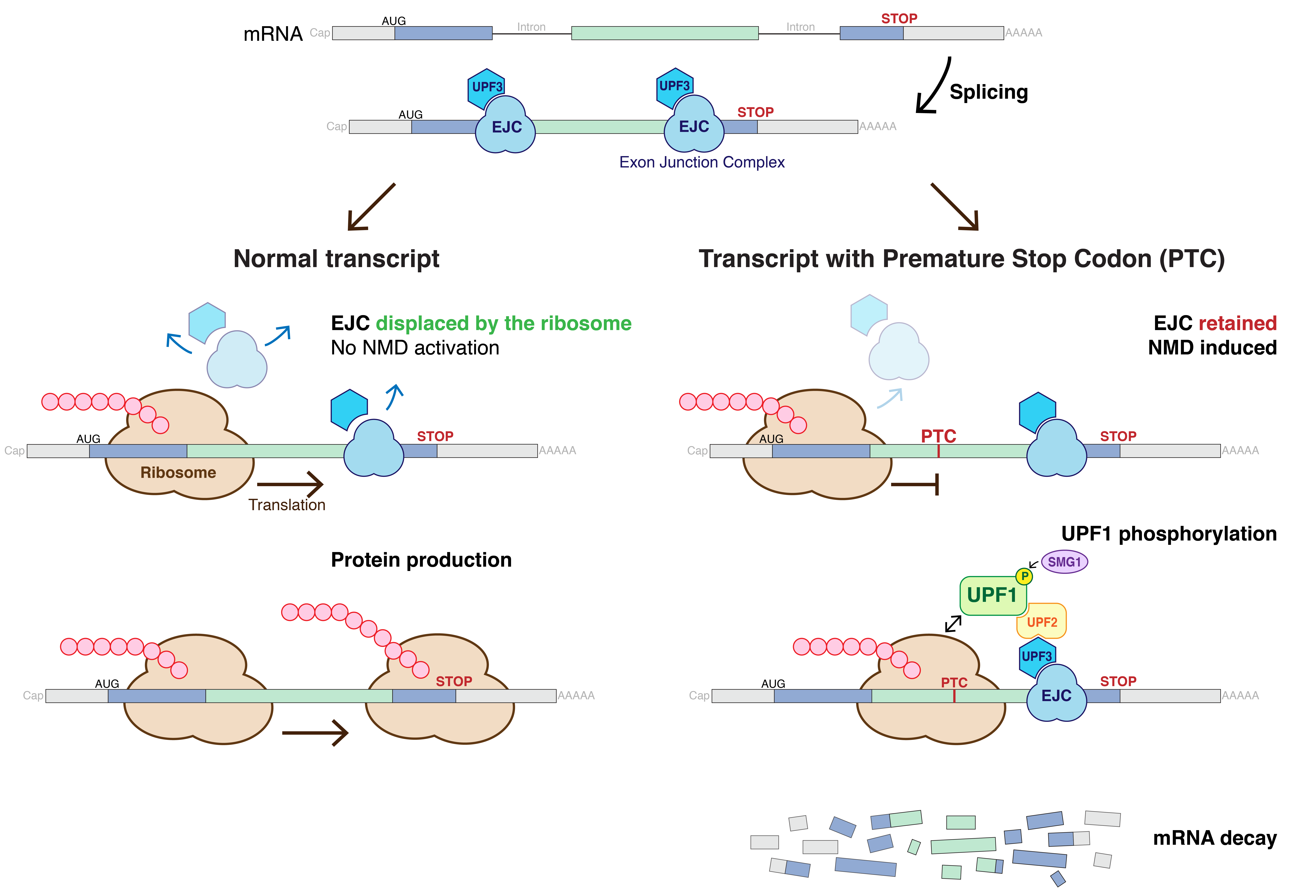
Nonsense-mediated Decay
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Translation of these aberrant mRNAs could, in some cases, lead to deleterious gain-of-function or dominant-negative activity of the resulting proteins. NMD was first described in human cells and in yeast almost simultaneously in 1979. This suggested broad phylogenetic conservation and an important biological role of this intriguing mechanism. NMD was discovered when it was realized that cells often contain unexpectedly low concentrations of mRNAs that are transcribed from alleles carrying nonsense mutations. Nonsense mutations code for a premature stop codon which causes the protein to be shortened. The truncated protein may or may not be functional, depending on the severity of what is not translated. In human genetics, NMD has the possibility to not only limit t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-induced Silencing Complex
The RNA-induced silencing complex, or RISC, is a multiprotein complex, specifically a ribonucleoprotein, which functions in gene silencing via a variety of pathways at the transcriptional and translational levels. Using single-stranded RNA (ssRNA) fragments, such as microRNA (miRNA), or double-stranded small interfering RNA (siRNA), the complex functions as a key tool in gene regulation. The single strand of RNA acts as a template for RISC to recognize complementary DNA, complementary messenger RNA (mRNA) transcription (genetics), transcript. Once found, one of the proteins in RISC, Argonaute, activates and cleaves the mRNA. This process is called RNA interference (RNAi) and it is found in many eukaryotes; it is a key process in defense against viral disease, viral infections, as it is triggered by the presence of double-stranded RNA (dsRNA). Discovery The biochemistry, biochemical identification of RISC was conducted by Gregory Hannon and his colleagues at the Cold Spring Harbor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MicroRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in messenger RNA (mRNA) molecules, then silence said mRNA molecules by one or more of the following processes: * Cleaving the mRNA strand into two pieces. * Destabilizing the mRNA by shortening its poly(A) tail. * Reducing translation of the mRNA into proteins. In cells of humans and other animals, miRNAs primarily act by destabilizing the mRNA. miRNAs resemble the small interfering RNAs (siRNAs) of the RNA interference (RNAi) pathway, except miRNAs derive from regions of RNA transcripts that fold back on themselves to form short stem-loops (hairpins), whereas siRNAs derive from longer regions of double-stranded RNA. The human genome may encode ov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA Decapping
The process of messenger RNA decapping consists of hydrolysis of the 5' cap structure on the RNA exposing a 5' monophosphate. In eukaryotes, this 5' monophosphate is a substrate for the 5' exonuclease Xrn1 and the mRNA is quickly destroyed. There are many situations which may lead to the removal of the cap, some of which are discussed below. In prokaryotes, the initial mRNA transcript naturally possesses a 5'-triphosphate group after bacterial transcription; the enzyme RppH removes a pyrophosphate molecule from the 5' end, converting the 5'-triphosphate to a 5'-monophosphate, triggering mRNA degradation by ribonucleases. Translation and decay Inside cells, there is a balance between the processes of translation and mRNA decay. Messages which are being actively translated are bound by polysomes and the eukaryotic initiation factors eIF-4E and eIF-4G (in eukaryotes). This blocks access to the cap by the decapping enzyme DCP2 and protects the mRNA molecule. In nutrient-starv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and the ribosome creates the protein utilizing amino acids carried by transfer RNA (tRNA). This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, genetic inf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Somatic Cells
In cellular biology, a somatic cell (), or vegetal cell, is any biological cell forming the body of a multicellular organism other than a gamete, germ cell, gametocyte or undifferentiated stem cell. Somatic cells compose the body of an organism and divide through mitosis. In contrast, gametes derive from meiosis within the germ cells of the germline and they fuse during sexual reproduction. Stem cells also can divide through mitosis, but are different from somatic in that they differentiate into diverse specialized cell types. In mammals, somatic cells make up all the internal organs, skin, bones, blood and connective tissue, while mammalian germ cells give rise to spermatozoa and ova which fuse during fertilization to produce a cell called a zygote, which divides and differentiates into the cells of an embryo. There are approximately 220 types of somatic cell in the human body. Theoretically, these cells are not germ cells (the source of gametes); they transmit their mutation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotic Cell
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of Outline of life forms, life forms alongside the two groups of prokaryotes: the Bacteria and the Archaea. Eukaryotes represent a small minority of the number of organisms, but given their generally much larger size, their collective global biomass is much larger than that of prokaryotes. The eukaryotes emerged within the archaeal Kingdom (biology), kingdom Asgard (Archaea), Promethearchaeati and its sole phylum Promethearchaeota. This implies that there are only Two-domain system, two domains of life, Bacteria and Archaea, with eukaryotes incorporated among the Archaea. Eukaryotes first emerged during the Paleoproterozoic, likely as Flagellated cell, flagellated cells. The leading evolutionary theory is they were created by symbiogenesis betwe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Streptavidin
Streptavidin is a 52 Atomic mass unit, kDa protein (tetramer) purified from the bacterium ''Streptomyces avidinii''. Streptavidin Homotetramer, homo-tetramers have an extraordinarily high affinity for biotin (also known as vitamin B7 or vitamin H). With a dissociation constant (Kd) on the order of ≈10−14 mol/L, the binding of biotin to streptavidin is one of the strongest non-covalent interactions known in nature. Streptavidin is used extensively in molecular biology and bionanotechnology due to the streptavidin-biotin complex's resistance to organic solvents, denaturants (e.g. guanidinium chloride), detergents (e.g. Sodium dodecyl sulfate, SDS, Triton X-100), proteolytic enzymes, and extremes of temperature and pH. Structure The crystal structure of streptavidin with biotin bound was reported by two groups in 1989. The structure was solved using multi wavelength anomalous diffraction by Hendrickson et al. at Columbia University and using multiple isomorphous replacemen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biotinylation
In biochemistry, biotinylation is the process of covalently attaching biotin to a protein, nucleic acid or other molecule. Biotinylation is rapid, specific and is unlikely to disturb the natural function of the molecule due to the small size of biotin (MW = 244.31 g/mol). Biotin binds to streptavidin and avidin with an extremely high affinity, fast on-rate, and high specificity, and these interactions are exploited in many areas of biotechnology to isolate biotinylated molecules of interest. Biotin-binding to streptavidin and avidin is resistant to extremes of heat, pH and proteolysis, making capture of biotinylated molecules possible in a wide variety of environments. Also, multiple biotin molecules can be Cross-link, conjugated to a protein of interest, which allows binding of multiple streptavidin, avidin or neutravidin protein molecules and increases the sensitivity of detection of the protein of interest. There is a large number of biotinylation reagents available that expl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biotin
Biotin (also known as vitamin B7 or vitamin H) is one of the B vitamins. It is involved in a wide range of metabolic processes, both in humans and in other organisms, primarily related to the utilization of fats, carbohydrates, and amino acids. The name ''biotin'', borrowed from the German , derives from the Ancient Greek word (; 'life') and the suffix "-in" (a suffix used in chemistry usually to indicate 'forming'). Biotin appears as a white, needle-like crystalline solid. Chemical description Biotin is classified as a heterocyclic compound, with a sulfur-containing tetrahydrothiophene ring fused to a ureido group. A C5-carboxylic acid side chain is appended to the former ring. The ureido ring, containing the −N−CO−N− group, serves as the carbon dioxide carrier in carboxylation reactions. Biotin is a coenzyme for five carboxylase enzymes, which are involved in the catabolism of amino acids and fatty acids, synthesis of fatty acids, and gluconeogenesis. Biotinylat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |