|
NhaA-II RNA Motif
The ''nhaA''-II RNA motif is a conserved RNA structure that was discovered by bioinformatics. ''nhaA''-II motifs are found in Caulobacterales. ''nhaA''-II motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes. ''nhaA''-I RNAs typically occur upstream of genes that encode exchangers of sodium ions and protons. More rarely, they also exist upstream of methyltransferases that use S-adenosylmethionine ''S''-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throug ... as a donor. References {{reflist Non-coding RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Conservation
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids (DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst eukaryotes, and the tmRNA in bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, promp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Element
''Cis''-regulatory elements (CREs) or ''cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics Discovery Of Non-coding RNAs
Non-coding RNAs have been discovered using both experimental and bioinformatic approaches. Bioinformatic approaches can be divided into three main categories. The first involves homology search, although these techniques are by definition unable to find new classes of ncRNAs. The second category includes algorithms designed to discover specific types of ncRNAs that have similar properties. Finally, some discovery methods are based on very general properties of RNA, and are thus able to discover entirely new kinds of ncRNAs. Discovery by homology search Homology search refers to the process of searching a sequence database for RNAs that are similar to already known RNA sequences. Any algorithm that is designed for homology search of nucleic acid sequences can be used, e.g., BLAST. However, such algorithms typically are not as sensitive or accurate as algorithms specifically designed for RNA. Of particular importance for RNA is its conservation of a secondary structure, which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Caulobacterales
''Caulobacterales'' is an order of gram-negative pseudomonadota Pseudomonadota (synonym "Proteobacteria") is a major phylum of gram-negative bacteria. Currently, they are considered the predominant phylum within the domain of bacteria. They are naturally found as pathogenic and free-living (non- parasitic) ... within the alpha subgroup.Garrity, George M.; Brenner, Don J.; Krieg, Noel R.; Staley, James T. (eds.) (2005). Bergey's Manual of Systematic Bacteriology, Volume Two: The Proteobacteria, Part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. New York, New York: Springer. . References Alphaproteobacteria Caulobacterales {{alphaproteobacteria-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Element
''Cis''-regulatory elements (CREs) or ''cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes. During gene expression (the synthesis of Gene product, RNA or protein from a gene), DNA is first transcription (biology), copied into RNA. RNA can be non-coding RNA, directly functional or be the intermediate protein biosynthesis, template for the synthesis of a protein. The transmission of genes to an organism's offspring, is the basis of the inheritance of phenotypic traits from one generation to the next. These genes make up different DNA sequences, together called a genotype, that is specific to every given individual, within the gene pool of the population (biology), population of a given species. The genotype, along with environmental and developmental factors, ultimately determines the phenotype ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
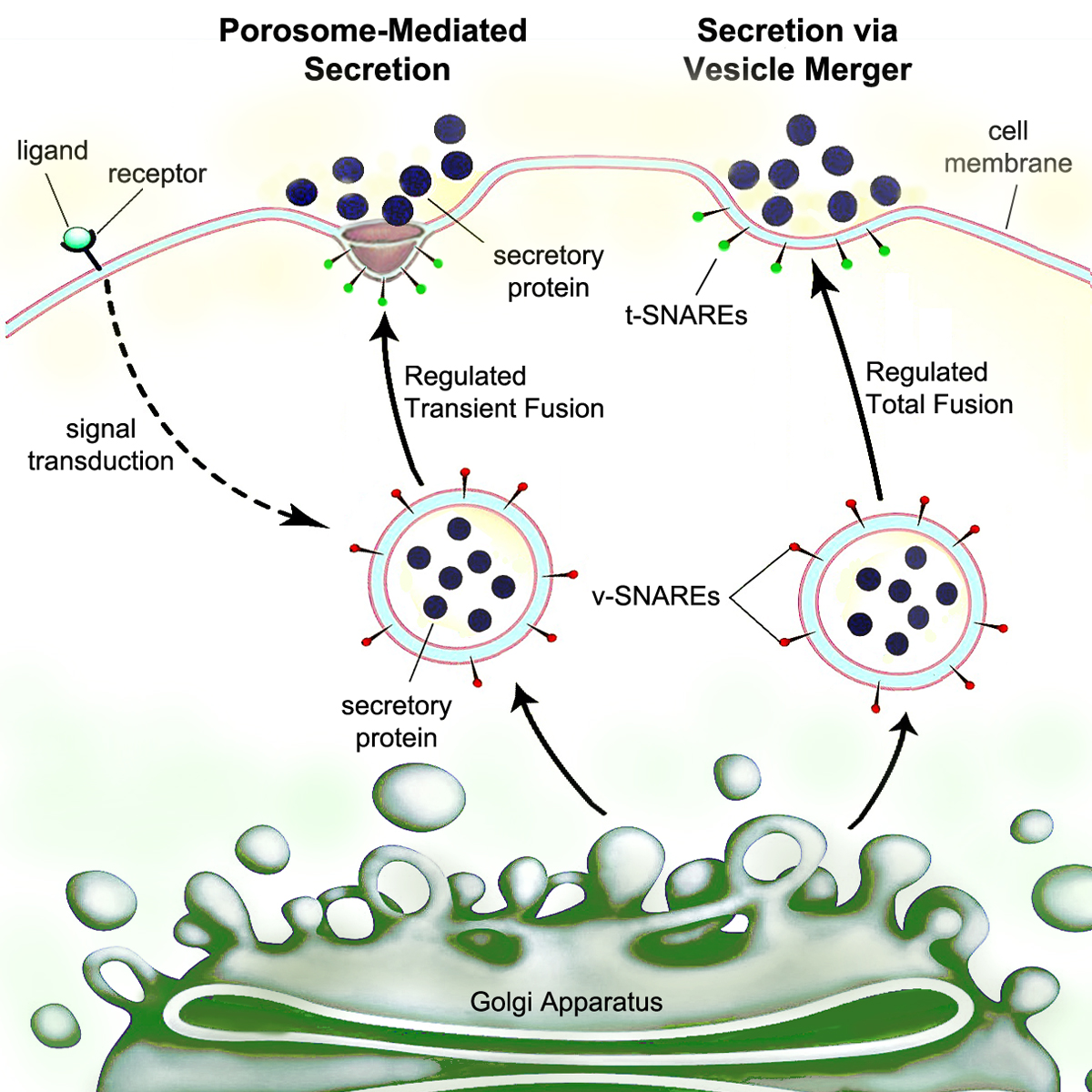
Secretion
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion is the removal of certain substances or waste products from a cell or organism. The classical mechanism of cell secretion is via secretory portals at the plasma membrane called porosomes. Porosomes are permanent cup-shaped lipoprotein structures embedded in the cell membrane, where secretory vesicles transiently dock and fuse to release intra-vesicular contents from the cell. Secretion in bacterial species means the transport or translocation of effector molecules. For example: proteins, enzymes or toxins (such as cholera toxin in pathogenic bacteria e.g. '' Vibrio cholerae'') from across the interior (cytoplasm or cytosol) of a bacterial cell to its exterior. Secretion is a very important mechanism in bacterial functioning and operation in their natural surrounding environment for adaptation and survival. In eukaryotic cells ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sodium
Sodium is a chemical element; it has Symbol (chemistry), symbol Na (from Neo-Latin ) and atomic number 11. It is a soft, silvery-white, highly reactive metal. Sodium is an alkali metal, being in group 1 element, group 1 of the periodic table. Its only stable isotope is 23Na. The free metal does not occur in nature and must be prepared from compounds. Sodium is the Abundance of elements in Earth's crust, sixth most abundant element in the Earth's crust and exists in numerous minerals such as feldspars, sodalite, and halite (NaCl). Many salts of sodium are highly water-soluble: sodium ions have been Leaching (chemistry), leached by the action of water from the Earth, Earth's minerals over eons, and thus sodium and chlorine are the most common dissolved elements by weight in the oceans. Sodium was first isolated by Humphry Davy in 1807 by the electrolysis of sodium hydroxide. Among many other useful sodium compounds, sodium hydroxide (lye) is used in Soap, soap manufac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ions
An ion () is an atom or molecule with a net electrical charge. The charge of an electron is considered to be negative by convention and this charge is equal and opposite to the charge of a proton, which is considered to be positive by convention. The net charge of an ion is not zero because its total number of electrons is unequal to its total number of protons. A cation is a positively charged ion with fewer electrons than protons (e.g. K+ (potassium ion)) while an anion is a negatively charged ion with more electrons than protons (e.g. Cl− (chloride ion) and OH− (hydroxide ion)). Opposite electric charges are pulled towards one another by electrostatic force, so cations and anions attract each other and readily form ionic compounds. Ions consisting of only a single atom are termed ''monatomic ions'', ''atomic ions'' or ''simple ions'', while ions consisting of two or more atoms are termed polyatomic ions or ''molecular ions''. If only a + or − is present, it indicates ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proton
A proton is a stable subatomic particle, symbol , Hydron (chemistry), H+, or 1H+ with a positive electric charge of +1 ''e'' (elementary charge). Its mass is slightly less than the mass of a neutron and approximately times the mass of an electron (the proton-to-electron mass ratio). Protons and neutrons, each with a mass of approximately one Dalton (unit), dalton, are jointly referred to as ''nucleons'' (particles present in atomic nuclei). One or more protons are present in the Atomic nucleus, nucleus of every atom. They provide the attractive electrostatic central force which binds the atomic electrons. The number of protons in the nucleus is the defining property of an element, and is referred to as the atomic number (represented by the symbol ''Z''). Since each chemical element, element is identified by the number of protons in its nucleus, each element has its own atomic number, which determines the number of atomic electrons and consequently the chemical characteristi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methyltransferase
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding ''S''-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |