|
Mutant Protein
A mutant protein is the protein product encoded by a gene with mutation. Mutated protein can have single amino acid change (minor, but still in many cases significant change leading to disease) or wide-range amino acid changes by e.g. truncation of C-terminus after introducing premature stop codon. See also * Site-directed mutagenesis * Phi value analysis * missense mutation * nonsense mutation * point mutation * frameshift mutation * silent mutation * single-nucleotide polymorphism In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a ... References Proteins Mutation {{protein-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metabolic reactions, DNA replication, Cell signaling, responding to stimuli, providing Cytoskeleton, structure to cells and Fibrous protein, organisms, and Intracellular transport, transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the Nucleic acid sequence, nucleotide sequence of their genes, and which usually results in protein folding into a specific Protein structure, 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called pep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes. During gene expression (the synthesis of Gene product, RNA or protein from a gene), DNA is first transcription (biology), copied into RNA. RNA can be non-coding RNA, directly functional or be the intermediate protein biosynthesis, template for the synthesis of a protein. The transmission of genes to an organism's offspring, is the basis of the inheritance of phenotypic traits from one generation to the next. These genes make up different DNA sequences, together called a genotype, that is specific to every given individual, within the gene pool of the population (biology), population of a given species. The genotype, along with environmental and developmental factors, ultimately determines the phenotype ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA (such as pyrimidine dimers caused by exposure to ultraviolet radiation), which then may undergo error-prone repair (especially microhomology-mediated end joining), cause an error during other forms of repair, or cause an error during replication ( translesion synthesis). Mutations may also result from substitution, insertion or deletion of segments of DNA due to mobile genetic elements. Mutations may or may not produce detectable changes in the observable characteristics ( phenotype) of an organism. Mutations play a part in both normal and abnormal biological processes including: evolution, cancer, and the development of the immune system, including junctional diversity. Mutati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Site-directed Mutagenesis
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional mutating changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenesis, it is used for investigating the structure and biological activity of DNA, RNA, and protein molecules, and for protein engineering. Site-directed mutagenesis is one of the most important laboratory techniques for creating DNA libraries by introducing mutations into DNA sequences. There are numerous methods for achieving site-directed mutagenesis, but with decreasing costs of oligonucleotide synthesis, artificial gene synthesis is now occasionally used as an alternative to site-directed mutagenesis. Since 2013, the development of the CRISPR/Cas9 technology, based on a prokaryotic viral defense system, has also allowed for the editing of the genome, and mutagenesis may be performed ''in vivo'' with relative ease. History Early atte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phi Value Analysis
Phi value analysis, \varphi analysis, or \varphi-value analysis is an experimental protein engineering technique for studying structures of the transition state and intermediates in protein folding and conformational changes. The structure of the folding transition state has to be found from kinetic measurements and is not accessible by equilibrium methods such as protein NMR or X-ray crystallography and intermediates are often mobile and partly unstructured by definition. In \varphi -value analysis, the folding kinetics and conformational folding stability of the wild-type protein are compared with those of point mutants to find ''phi values''. These measure the mutant residue's energetic contribution to the folding transition state, which reveals the degree of native structure around the mutated residue in the transition state, by accounting for the relative free energies of the unfolded state, the folded state, and the transition state for the wild-type and mutant proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
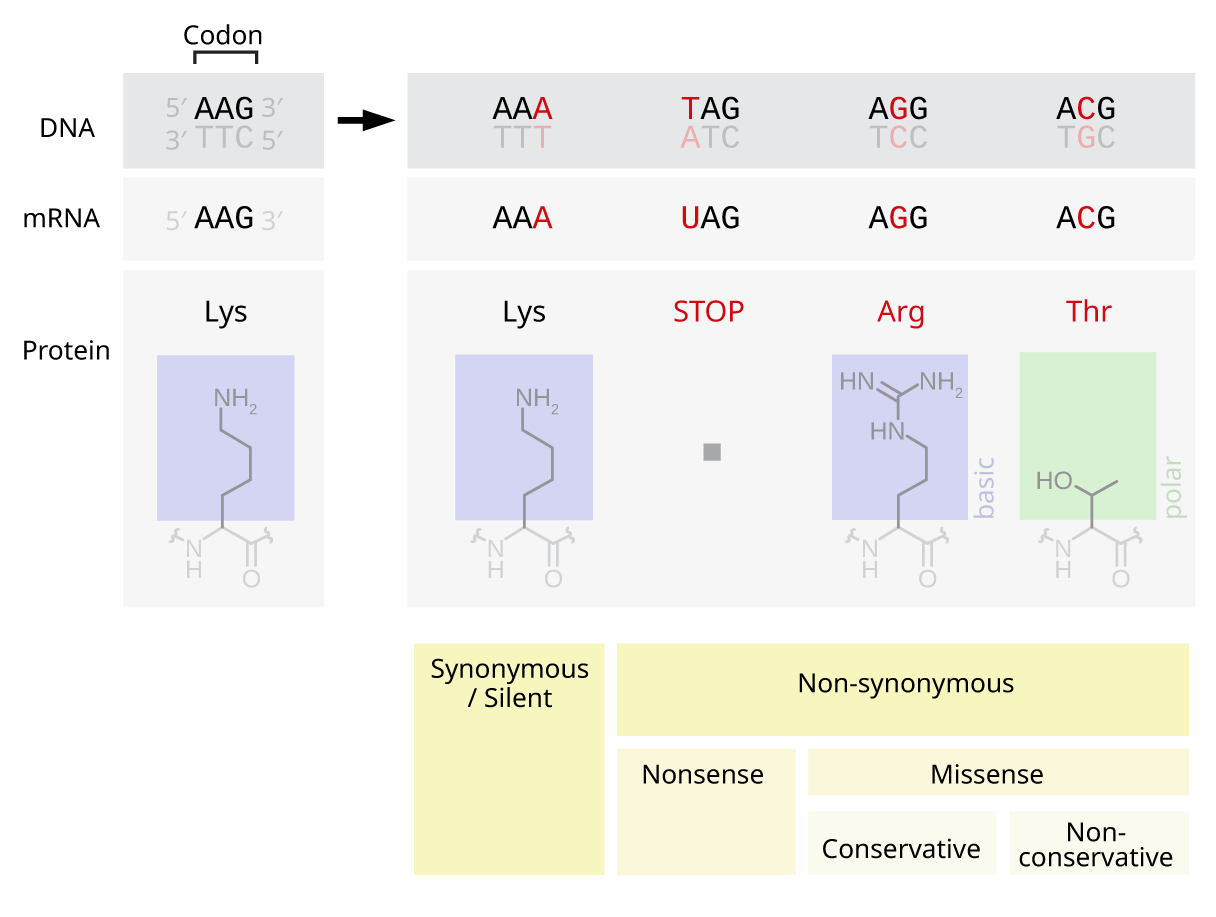
Missense Mutation
In genetics, a missense mutation is a point mutation in which a single nucleotide change results in a codon that codes for a different amino acid. It is a type of nonsynonymous substitution. Missense mutations change amino acids, which in turn alter proteins and may alter a protein's function or structure. These mutations may arise spontaneously from mutagens like UV radiation, tobacco smoke, an error in DNA replication, and other factors. Screening for missense mutations can be done by sequencing the genome of an organism and comparing the sequence to a reference genome to analyze for differences. Missense mutations can be repaired by the cell when there are errors in DNA replication by using mechanisms such as DNA proofreading and DNA mismatch repair, mismatch repair. They can also be repaired by using genetic engineering technologies or pharmaceuticals. Some notable examples of human diseases caused by missense mutations are Rett syndrome, cystic fibrosis, and Sickle cell disease ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
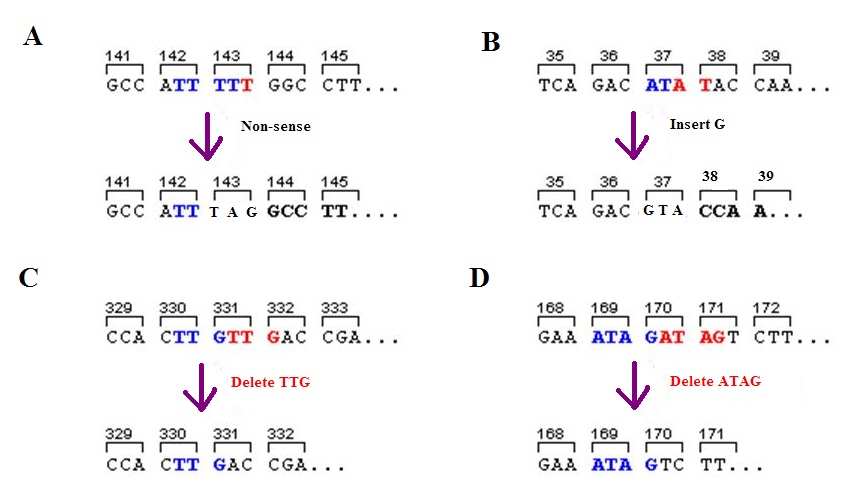
Nonsense Mutation
In genetics, a nonsense mutation is a point mutation in a sequence of DNA that results in a ''nonsense codon'', or a premature stop codon in the transcribed mRNA, and leads to a truncated, incomplete, and possibly nonfunctional protein product. Nonsense mutations are not always harmful; the functional effect of a nonsense mutation depends on many aspects, such as the location of the stop codon within the coding DNA. For example, the effect of a nonsense mutation depends on the proximity of the nonsense mutation to the original stop codon, and the degree to which functional subdomains of the protein are affected. As nonsense mutations leads to premature termination of polypeptide chains; they are also called chain termination mutations. Missense mutations differ from nonsense mutations since they are point mutations that exhibit a single nucleotide change to cause substitution of a different amino acid. A nonsense mutation also differs from a nonstop mutation, which is a point muta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Point Mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g. Synonymous substitution, synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function. Causes Point mutations usually take place during DNA replication. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one purine or pyrimidine may change the amino acid that the nucleotides code for. Point mutations may arise from spontaneous mutations that occur during DNA re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Frameshift Mutation
A frameshift mutation (also called a framing error or a reading frame shift) is a genetic mutation caused by indels ( insertions or deletions) of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet nature of gene expression by codons, the insertion or deletion can change the reading frame (the grouping of the codons), resulting in a completely different translation from the original. The earlier in the sequence the deletion or insertion occurs, the more altered the protein. A frameshift mutation is not the same as a single-nucleotide polymorphism in which a nucleotide is replaced, rather than inserted or deleted. A frameshift mutation will in general cause the reading of the codons after the mutation to code for different amino acids. The frameshift mutation will also alter the first stop codon ("UAA", "UGA" or "UAG") encountered in the sequence. The polypeptide being created could be abnormally short or abnormally long, and will most li ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Silent Mutation
Silent mutations, also called synonymous or samesense mutations, are mutations in DNA that do not have an observable effect on the organism's phenotype. The phrase ''silent mutation'' is often used interchangeably with the phrase '' synonymous mutation''; however, synonymous mutations are not always silent, nor vice versa. Synonymous mutations can affect transcription, splicing, mRNA transport, and translation, any of which could alter phenotype, rendering the synonymous mutation non-silent. The substrate specificity of the tRNA to the rare codon can affect the timing of translation, and in turn the co-translational folding of the protein. This is reflected in the codon usage bias that is observed in many species. Mutations that cause the altered codon to produce an amino acid with similar functionality (''e.g.'' a mutation producing leucine instead of isoleucine) are often classified as silent; if the properties of the amino acid are conserved, this mutation does not usually s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, a Guanine, G nucleotide present at a specific location in a reference genome may be replaced by an Adenine, A in a minority of individuals. The two possible nucleotide variations of this SNP – G or A – are called alleles. SNPs can help explain differences in susceptibility to a wide range of diseases across a population. For example, a common SNP in the Factor H, CFH gene is associated with increased risk of age-related macular degeneration. Differences in the severity of an illness or response to treatments may also be manifestations of genetic variations caused by SNPs. For example, two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Footnotes
In publishing, a note is a brief text in which the author comments on the subject and themes of the book and names supporting citations. In the editorial production of books and documents, typographically, a note is usually several lines of text at the bottom of the page, at the end of a chapter, at the end of a volume, or a house-style typographic usage throughout the text. Notes are usually identified with superscript numbers or a symbol.''The Oxford Companion to the English Language'' (1992) p. 709. Footnotes are informational notes located at the foot of the thematically relevant page, whilst endnotes are informational notes published at the end of a chapter, the end of a volume, or the conclusion of a multi-volume book. Unlike footnotes, which require manipulating the page design (text-block and page layouts) to accommodate the additional text, endnotes are advantageous to editorial production because the textual inclusion does not alter the design of the publication. H ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |