|
Monophyletic Groups
In biological cladistics for the classification of organisms, monophyly is the condition of a taxonomic grouping being a clade – that is, a grouping of organisms which meets these criteria: # the grouping contains its own most recent common ancestor (or more precisely an ancestral population), i.e. excludes non-descendants of that common ancestor # the grouping contains all the descendants of that common ancestor, without exception Monophyly is contrasted with paraphyly and polyphyly as shown in the second diagram. A ''paraphyletic'' grouping meets 1. but not 2., thus consisting of the descendants of a common ancestor, excepting one or more monophyletic subgroups. A ''polyphyletic'' grouping meets neither criterion, and instead serves to characterize convergent relationships of biological features rather than genetic relationships – for example, night-active primates, fruit trees, or aquatic insects. As such, these characteristic features of a polyphyletic grouping are n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Systematics
Systematics is the study of the diversification of living forms, both past and present, and the relationships among living things through time. Relationships are visualized as evolutionary trees (synonyms: phylogenetic trees, phylogenies). Phylogenies have two components: branching order (showing group relationships, graphically represented in cladograms) and branch length (showing amount of evolution). Phylogenetic trees of species and higher taxa are used to study the evolution of traits (e.g., anatomical or molecular characteristics) and the distribution of organisms ( biogeography). Systematics, in other words, is used to understand the evolutionary history of life on Earth. The word systematics is derived from the Latin word of Ancient Greek origin '' systema,'' which means systematic arrangement of organisms. Carl Linnaeus used 'Systema Naturae' as the title of his book. Branches and applications In the study of biological systematics, researchers use the different br ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paraphyly
Paraphyly is a taxonomic term describing a grouping that consists of the grouping's last common ancestor and some but not all of its descendant lineages. The grouping is said to be paraphyletic ''with respect to'' the excluded subgroups. In contrast, a monophyletic grouping (a clade) includes a common ancestor and ''all'' of its descendants. The terms are commonly used in phylogenetics (a subfield of biology) and in the tree model of historical linguistics. Paraphyletic groups are identified by a combination of synapomorphies and symplesiomorphies. If many subgroups are missing from the named group, it is said to be polyparaphyletic. The term received currency during the debates of the 1960s and 1970s accompanying the rise of cladistics, having been coined by zoologist Willi Hennig to apply to well-known taxa like Reptilia (reptiles), which is paraphyletic with respect to birds. Reptilia contains the last common ancestor of reptiles and all descendants of that ancest ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monotypic Taxon
In biology, a monotypic taxon is a taxonomic group (taxon) that contains only one immediately subordinate taxon. A monotypic species is one that does not include subspecies or smaller, infraspecific taxa. In the case of Genus, genera, the term "unispecific" or "monospecific" is sometimes preferred. In botanical nomenclature, a monotypic genus is a genus in the special case where a genus and a single species are simultaneously described. Theoretical implications Monotypic taxa present several important theoretical challenges in biological classification. One key issue is known as "Gregg's Paradox": if a single species is the only member of multiple hierarchical levels (for example, being the only species in its genus, which is the only genus in its family), then each level needs a distinct definition to maintain logical structure. Otherwise, the different taxonomic ranks become effectively identical, which creates problems for organizing biological diversity in a hierarchical o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glossary Of Scientific Naming
This is a list of terms and symbols used in scientific names for organisms, and in describing the names. For proper parts of the names themselves, see List of Latin and Greek words commonly used in systematic names. Many of the abbreviations are used with or without a full stop, stop. Naming standards and taxonomic organizations and their codes and taxonomies * ICTV – International Committee on Taxonomy of Viruses * ICSP – International Committee on Systematics of Prokaryotes ** formerly the ICSB – International Committee on Systematic Bacteriology ** publishes the ICNP – International Code of Nomenclature of Prokaryotes *** formerly the International Code of Nomenclature of Bacteria (ICNB) or Bacteriological Code (BC) * ICZN – International Commission on Zoological Nomenclature ** publishes ''ICZN'' – the ''International Code of Zoological Nomenclature'' or "ICZN Code" * IBC – International Botanical Congress ** publishes ''ICN'' – the ''International Code of Nome ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Crown Group
In phylogenetics, the crown group or crown assemblage is a collection of species composed of the living representatives of the collection, the most recent common ancestor of the collection, and all descendants of the most recent common ancestor. It is thus a way of defining a clade, a group consisting of a species and all its extant or extinct descendants. For example, Neornithes (birds) can be defined as a crown group, which includes the most recent common ancestor of all modern birds, and all of its extant or extinct descendants. The concept was developed by Willi Hennig, the formulator of phylogenetic systematics, as a way of classifying living organisms relative to their extinct relatives in his "Die Stammesgeschichte der Insekten", and the "crown" and "stem" group terminology was coined by R. P. S. Jefferies in 1979. Though formulated in the 1970s, the term was not commonly used until its reintroduction in 2000 by Graham Budd and Sören Jensen. Contents of the crow ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Clade
In biology, a clade (), also known as a Monophyly, monophyletic group or natural group, is a group of organisms that is composed of a common ancestor and all of its descendants. Clades are the fundamental unit of cladistics, a modern approach to taxonomy adopted by most biological fields. The common ancestor may be an individual, a population, or a species (extinct or Extant taxon, extant). Clades are nested, one in another, as each branch in turn splits into smaller branches. These splits reflect evolutionary history as populations diverged and evolved independently. Clades are termed ''monophyletic'' (Greek: "one clan") groups. Over the last few decades, the cladistic approach has revolutionized biological classification and revealed surprising evolutionary relationships among organisms. Increasingly, taxonomists try to avoid naming Taxon, taxa that are not clades; that is, taxa that are not Monophyly, monophyletic. Some of the relationships between organisms that the molecul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
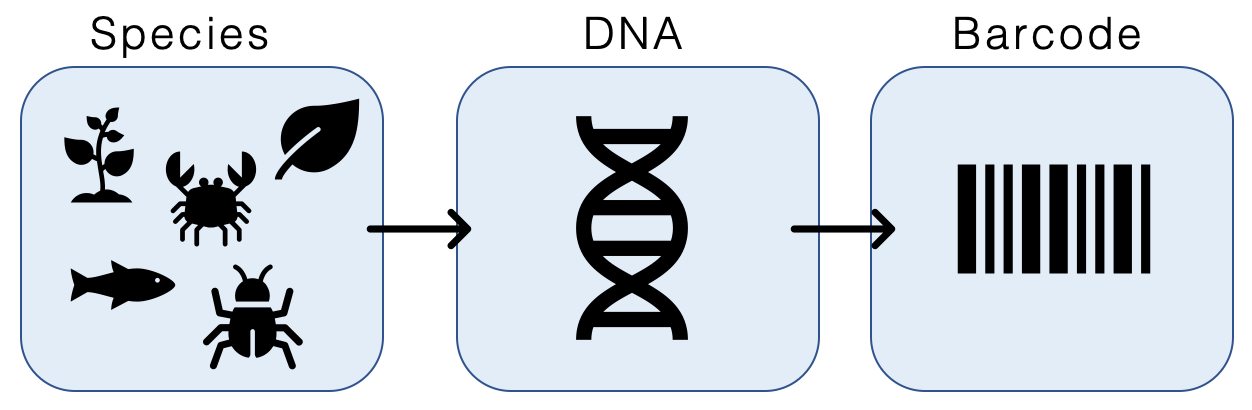
DNA Barcoding
DNA barcoding is a method of species identification using a short section of DNA from a specific gene or genes. The premise of DNA barcoding is that by comparison with a reference library of such DNA sections (also called " sequences"), an individual sequence can be used to uniquely identify an organism to species, just as a supermarket scanner uses the familiar black stripes of the UPC barcode to identify an item in its stock against its reference database. These "barcodes" are sometimes used in an effort to identify unknown species or parts of an organism, simply to catalog as many taxa as possible, or to compare with traditional taxonomy in an effort to determine species boundaries. Different gene regions are used to identify the different organismal groups using barcoding. The most commonly used barcode region for animals and some protists is a portion of the cytochrome ''c'' oxidase I (COI or COX1) gene, found in mitochondrial DNA. Other genes suitable for DNA barcoding a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hybrid Speciation
Hybrid speciation is a form of speciation where hybridization between two different species leads to a new species, reproductively isolated from the parent species. Previously, reproductive isolation between two species and their parents was thought to be particularly difficult to achieve, and thus hybrid species were thought to be very rare. With DNA analysis becoming more accessible in the 1990s, hybrid speciation has been shown to be a somewhat common phenomenon, particularly in plants. In botanical nomenclature, a hybrid species is also called a nothospecies. Hybrid species are by their nature polyphyletic. Ecology A hybrid may occasionally be better fitted to the local environment than the parental lineage, and as such, natural selection may favor these individuals. If reproductive isolation is subsequently achieved, a separate species may arise. Reproductive isolation may be genetic, ecological, behavioral, spatial, or a combination of these. If reproductive isolation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Convergent Evolution
Convergent evolution is the independent evolution of similar features in species of different periods or epochs in time. Convergent evolution creates analogous structures that have similar form or function but were not present in the last common ancestor of those groups. The cladistic term for the same phenomenon is Cladogram#Homoplasies, homoplasy. The recurrent evolution of flight is a classic example, as flying pterygota, insects, birds, pterosaurs, and bats have independently evolved the useful capacity of flight. Functionally similar features that have arisen through convergent evolution are ''analogous'', whereas ''homology (biology), homologous'' structures or traits have a common origin but can have dissimilar functions. Bird, bat, and pterosaur wings are analogous structures, but their forelimbs are homologous, sharing an ancestral state despite serving different functions. The opposite of convergence is divergent evolution, where related species evolve different trai ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Symplesiomorphy
In phylogenetics, a plesiomorphy ("near form") and symplesiomorphy are synonyms for an ancestral character shared by all members of a clade, which does not distinguish the clade from other clades. Plesiomorphy, symplesiomorphy, apomorphy, and synapomorphy all mean a trait shared between species because they share an ancestral species. Apomorphic and synapomorphic characteristics convey much information about evolutionary clades and can be used to define taxa. However, plesiomorphic and symplesiomorphic characteristics cannot. The term ''symplesiomorphy'' was introduced in 1950 by German entomologist Willi Hennig. Examples A backbone is a plesiomorphic trait shared by birds and mammals, and does not help in placing an animal in one or the other of these two clades. Birds and mammals share this trait because both clades are descended from the same far distant ancestor. Other clades, e.g. snakes, lizards, turtles, fish, frogs, all have backbones and none are either birds n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Synapomorphy
In phylogenetics, an apomorphy (or derived trait) is a novel Phenotypic trait, character or character state that has evolution, evolved from its ancestral form (or Plesiomorphy and symplesiomorphy, plesiomorphy). A synapomorphy is an apomorphy shared by two or more taxon, taxa and is therefore Hypothesis#Scientific hypothesis, hypothesized to have evolved in their most recent common ancestor. ) In cladistics, synapomorphy implies Homology (biology), homology. Examples of apomorphy are the presence of Terrestrial locomotion#Posture, erect gait, fur, Evolution of mammalian auditory ossicles, the evolution of three middle ear bones, and mammary glands in mammals but not in other vertebrate animals such as amphibians or reptiles, which have retained their ancestral traits of a Terrestrial locomotion#Posture, sprawling gait and lack of fur. Thus, these derived traits are also synapomorphies of mammals in general as they are not shared by other vertebrate animals. Etymology The word ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |