|
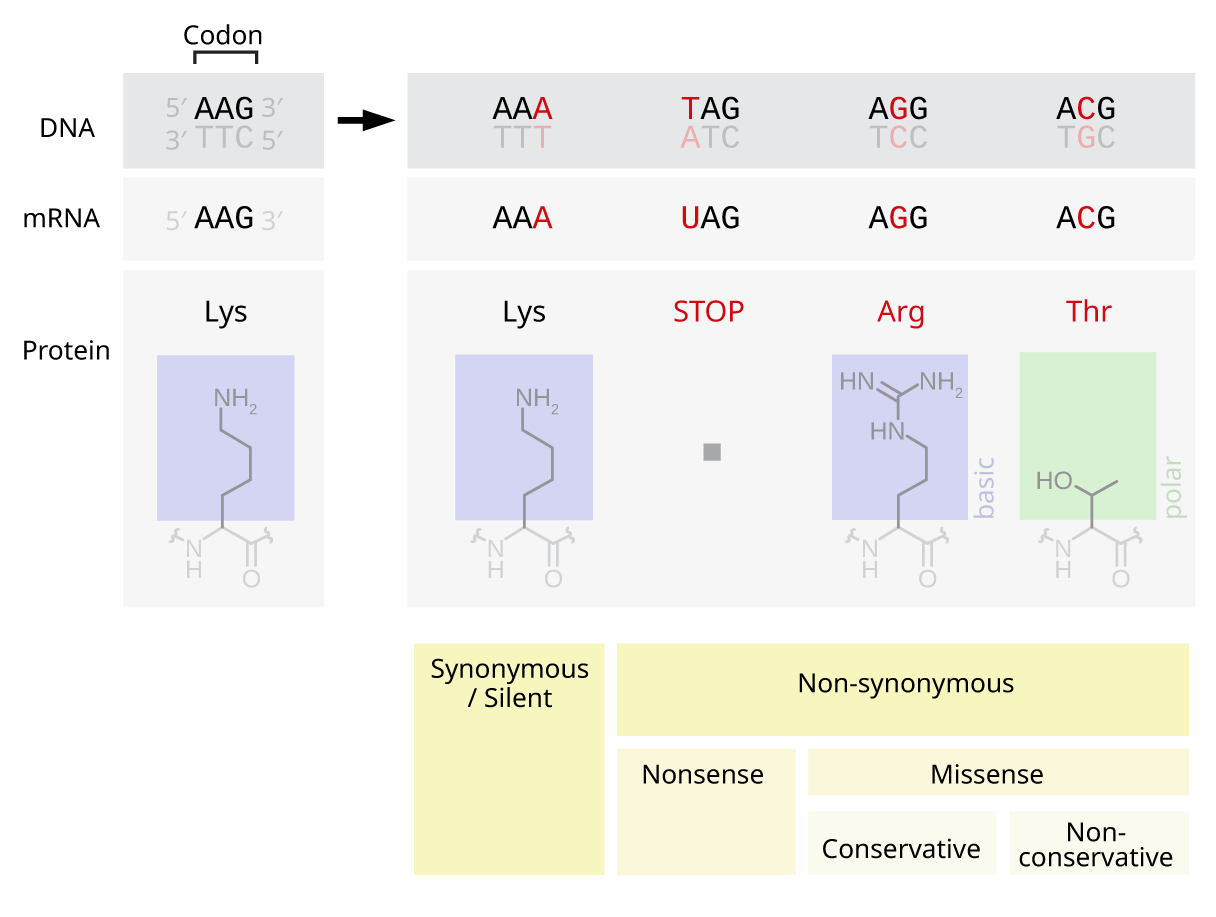
Missense
In genetics, a missense mutation is a point mutation in which a single nucleotide change results in a codon that codes for a different amino acid. It is a type of nonsynonymous substitution. Missense mutations change amino acids, which in turn alter proteins and may alter a protein's function or structure. These mutations may arise spontaneously from mutagens like UV radiation, tobacco smoke, an error in DNA replication, and other factors. Screening for missense mutations can be done by sequencing the genome of an organism and comparing the sequence to a reference genome to analyze for differences. Missense mutations can be repaired by the cell when there are errors in DNA replication by using mechanisms such as DNA proofreading and mismatch repair. They can also be repaired by using genetic engineering technologies or pharmaceuticals. Some notable examples of human diseases caused by missense mutations are Rett syndrome, cystic fibrosis, and sickle-cell disease. Impact on Protei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Missense Mutation Example
In genetics, a missense mutation is a point mutation in which a single nucleotide change results in a codon that codes for a different amino acid. It is a type of nonsynonymous substitution. Missense mutations change amino acids, which in turn alter proteins and may alter a protein's function or structure. These mutations may arise spontaneously from mutagens like UV radiation, tobacco smoke, an error in DNA replication, and other factors. Screening for missense mutations can be done by sequencing the genome of an organism and comparing the sequence to a reference genome to analyze for differences. Missense mutations can be repaired by the cell when there are errors in DNA replication by using mechanisms such as DNA proofreading and mismatch repair. They can also be repaired by using genetic engineering technologies or pharmaceuticals. Some notable examples of human diseases caused by missense mutations are Rett syndrome, cystic fibrosis, and sickle-cell disease. Impact on Protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Point Mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g. Synonymous substitution, synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function. Causes Point mutations usually take place during DNA replication. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one purine or pyrimidine may change the amino acid that the nucleotides code for. Point mutations may arise from spontaneous mutations that occur during DNA re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
De Novo Mutation
A de novo mutation (DNM) is any mutation or alteration in the genome of an individual organism (human, animal, plant, microbe, etc.) that was not inherited from its parents. This type of mutation spontaneously occurs during the process of DNA replication during cell division. De novo mutations, by definition, are present in the affected individual but absent from both biological parents' genomes. A de novo mutation can arise in a sperm or egg cell and become a germline mutation, or after fertilization as a post-zygotic mutation that cannot be inherited by offspring. These mutations can occur in any cell of the offspring, but those in the germ line (eggs or sperm) can be passed on to the next generation. In most cases, such a mutation has little or no effect on the affected organism due to the redundancy and robustness of the genetic code. However, in rare cases, it can have notable and serious effects on overall health, physical appearance, and other traits. Disorders that most co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stop Codon
In molecular biology, a stop codon (or termination codon) is a codon (nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain. While start codons need nearby sequences or initiation factors to start translation, a stop codon alone is sufficient to initiate termination. Properties Standard codons In the standard genetic code, there are three different termination codons: Alternative stop codons There are variations on the standard genetic code, and alternative stop codons have been found in the mitochondrial genomes of vertebrates, '' Scenedesmus obliquus'', and '' Thraustochytrium''. Reassigned ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Silent Mutation
Silent mutations, also called synonymous or samesense mutations, are mutations in DNA that do not have an observable effect on the organism's phenotype. The phrase ''silent mutation'' is often used interchangeably with the phrase '' synonymous mutation''; however, synonymous mutations are not always silent, nor vice versa. Synonymous mutations can affect transcription, splicing, mRNA transport, and translation, any of which could alter phenotype, rendering the synonymous mutation non-silent. The substrate specificity of the tRNA to the rare codon can affect the timing of translation, and in turn the co-translational folding of the protein. This is reflected in the codon usage bias that is observed in many species. Mutations that cause the altered codon to produce an amino acid with similar functionality (''e.g.'' a mutation producing leucine instead of isoleucine) are often classified as silent; if the properties of the amino acid are conserved, this mutation does not usually s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinians, Augustinian friar working in the 19th century in Brno, was the first to study genetics scientifically. Mendel studied "trait inheritance", patterns in the way traits are handed down from parents to offspring over time. He observed that organisms (pea plants) inherit traits by way of discrete "units of inheritance". This term, still used today, is a somewhat ambiguous definition of what is referred to as a gene. Phenotypic trait, Trait inheritance and Molecular genetics, molecular inheritance mechanisms of genes are still primary principles of genetics in the 21st century, but modern genetics has expanded to study the function and behavior of genes. Gene structure and function, variation, and distribution are studied within the context of the Cell (bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Codon
Genetic code is a set of rules used by living cells to translate information encoded within genetic material (DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries. The codons specify which amino acid will be added next during protein biosynthesis. With some exceptions, a three-nucleotide codon in a nucleic acid sequence specifies a single amino acid. The vast majority of genes are encoded with a single scheme (see the RNA codon table). That scheme is often called the canonical or standard genetic code, or simply ''the'' genetic code, though variant codes (such as in mitochondria) exist. History Efforts to understan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reference Genome
A reference genome (also known as a reference assembly) is a digital nucleic acid sequence database, assembled by scientists as a representative example of the genome, set of genes in one idealized individual organism of a species. As they are assembled from the sequencing of DNA from a number of individual donors, reference genomes do not accurately represent the set of genes of any single individual organism. Instead, a reference provides a haploid mosaic of different DNA sequences from each donor. For example, one of the most recent human reference genomes, assembly ''GRCh38, GRCh38/hg38'', is derived from >60 genomic library, genomic clone libraries. There are reference genomes for multiple species of viruses, bacteria, fungus, plants, and animals. Reference genomes are typically used as a guide on which new genomes are built, enabling them to be assembled much more quickly and cheaply than the initial Human Genome Project. Reference genomes can be accessed online at several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Superoxide Dismutase
Superoxide dismutase (SOD, ) is an enzyme that alternately catalyzes the dismutation (or partitioning) of the superoxide () anion radical into normal molecular oxygen (O2) and hydrogen peroxide (). Superoxide is produced as a by-product of oxygen metabolism and, if not regulated, causes many types of cell damage. Hydrogen peroxide is also damaging and is degraded by other enzymes such as catalase. Thus, SOD is an important antioxidant defense in nearly all living cells exposed to oxygen. One exception is '' Lactobacillus plantarum'' and related lactobacilli, which use intracellular manganese to prevent damage from reactive . Chemical reaction SODs catalyze the disproportionation of superoxide: : + → + In this way, is converted into two less damaging species. The general form, applicable to all the different metal−coordinated forms of SOD, can be written as follows: * + → + * + + → + The reactions by which SOD−catalyzed dismutation of superoxide for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
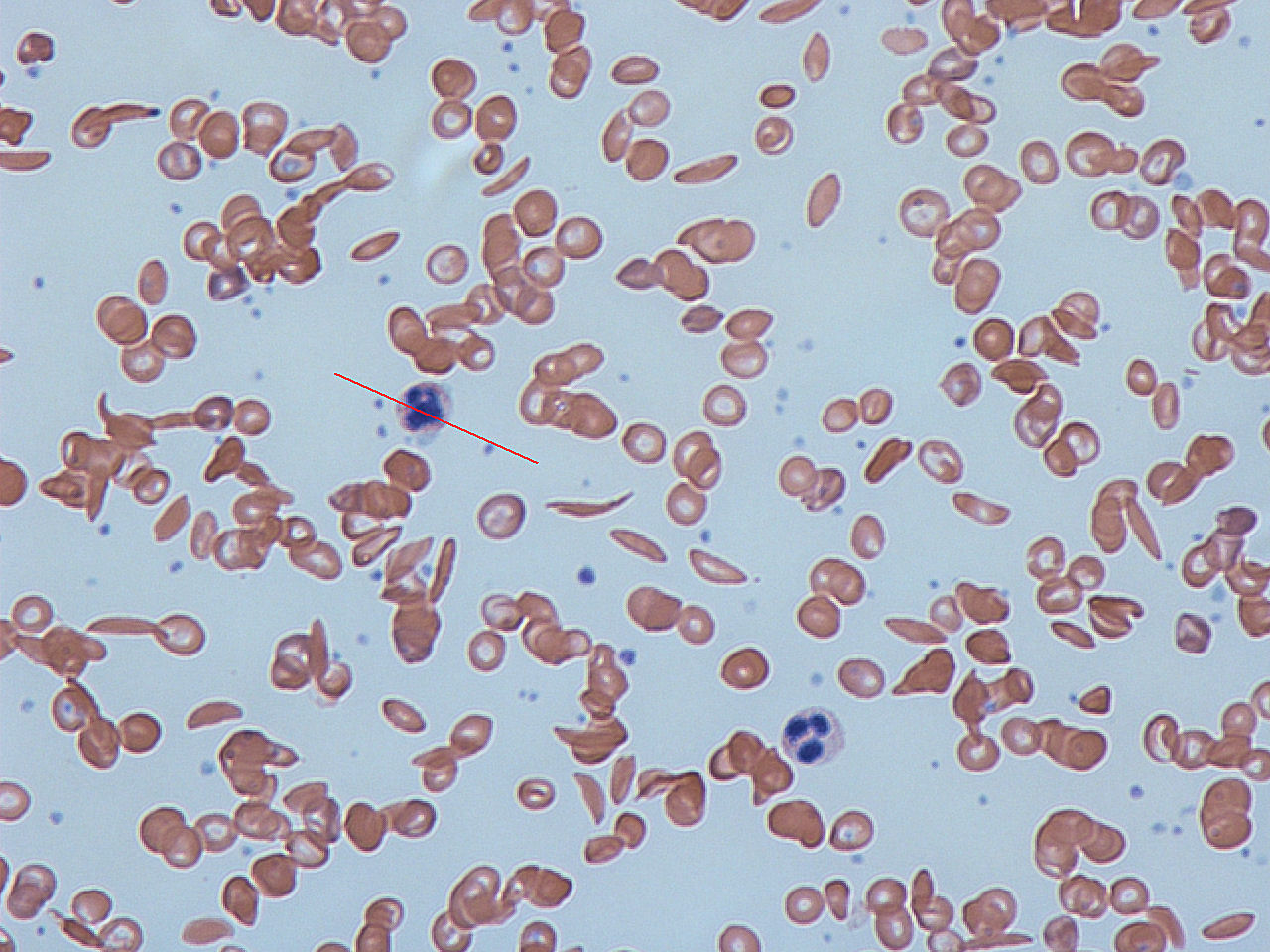
Newborn Screening
Newborn screening (NBS) is a public health program of screening (medicine), screening in infants shortly after birth for conditions that are treatable, but not clinically evident in the newborn period. The goal is to identify infants at risk for these conditions early enough to confirm the diagnosis and provide intervention that will alter the clinical course of the disease and prevent or ameliorate the clinical manifestations. NBS started with the discovery that the amino acid disorder phenylketonuria (PKU) could be treated by dietary adjustment, and that early intervention was required for the best outcome. Infants with PKU appear normal at birth, but are unable to metabolize the essential amino acid phenylalanine, resulting in irreversible intellectual disability. In the 1960s, Robert Guthrie (microbiologist), Robert Guthrie developed a simple method using a bacterial inhibition assay that could detect high levels of phenylalanine in blood shortly after a baby was born. Guth ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Massive Parallel Sequencing
Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of massively parallel processing; it is also called next-generation sequencing (NGS) or second-generation sequencing. Some of these technologies emerged between 1993 and 1998 and have been commercially available since 2005. These technologies use miniaturized and parallelized platforms for sequencing of 1 million to 43 billion short reads (50 to 400 bases each) per instrument run. Many NGS platforms differ in engineering configurations and sequencing chemistry. They share the technical paradigm of massive parallel sequencing via spatially separated, clonally amplified DNA templates or single DNA molecules in a flow cell. This design is very different from that of Sanger sequencing—also known as capillary sequencing or first-generation sequencing—which is based on electrophoretic separation of chain-termination products produced in ind ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |