|
Minutiae
A fingerprint is an impression left by the friction ridges of a human finger. The recovery of partial fingerprints from a crime scene is an important method of forensic science. Moisture and grease on a finger result in fingerprints on surfaces such as glass or metal. Deliberate impressions of entire fingerprints can be obtained by ink or other substances transferred from the peaks of friction ridges on the skin to a smooth surface such as paper. Fingerprint records normally contain impressions from the pad on the last joint of fingers and thumbs, though fingerprint cards also typically record portions of lower joint areas of the fingers. Human fingerprints are detailed, unique, difficult to alter, and durable over the life of an individual, making them suitable as long-term markers of human identity. They may be employed by police or other authorities to identify individuals who wish to conceal their identity, or to identify people who are incapacitated or dead and thus unab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biometric
Biometrics are body measurements and calculations related to human characteristics and features. Biometric authentication (or realistic authentication) is used in computer science as a form of identification and access control. It is also used to identify individuals in groups that are under surveillance. Biometric identifiers are the distinctive, measurable characteristics used to label and describe individuals. Biometric identifiers are often categorized as physiological characteristics which are related to the shape of the body. Examples include, but are not limited to fingerprint, palm veins, face recognition, DNA, palm print, hand geometry, iris recognition, retina, odor/scent, voice, shape of ears and gait. Behavioral characteristics are related to the pattern of behavior of a person, including but not limited to mouse movement, typing rhythm, gait, signature, voice, and behavioral profiling. Some researchers have coined the term behaviometrics (behavioral biom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Additive Genetic Effects
Genetic effects are broadly divided into two categories: additive and non-additive. Additive genetic effects occur where expression of more than one gene contributes to phenotype (or where alleles of a heterozygous gene both contribute), and the phenotypic expression of these gene(s) can be said to be the sum of these contributions. Non-additive effects involve dominance or epistasis, and cause outcomes that are not a sum of the contribution of the genes involved. Additive genetic effects are singularly important with regard to quantitative traits, as the sum of these effects informs the placement of a trait on the spectrum of possible outcomes. Quantitative traits are commonly polygenic (resulting from the effects of more than one locus). Heritability Broad sense heritability Variation in phenotypes across a population arises from the interaction between environmental variation and genetic variation between individuals. This can be stated mathematically as: VP = VE + ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antisense RNA
Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its Translation (biology), translation into protein. The asRNAs (which occur naturally) have been found in both prokaryotes and eukaryotes, and can be classified into short (200 nucleotides) non-coding RNAs (ncRNAs). The primary function of asRNA is regulating gene expression. asRNAs may also be produced synthetically and have found wide spread use as research tools for gene knockdown. They may also have therapeutic applications. Discovery and history in drug development Some of the earliest asRNAs were discovered while investigating functional proteins. An example was MicF RNA, micF asRNA. While characterizing the outer membrane Porin (protein), porin in Escherichia coli, ''E.coli'', some of the promoter clones observ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, a Guanine, G nucleotide present at a specific location in a reference genome may be replaced by an Adenine, A in a minority of individuals. The two possible nucleotide variations of this SNP – G or A – are called alleles. SNPs can help explain differences in susceptibility to a wide range of diseases across a population. For example, a common SNP in the Factor H, CFH gene is associated with increased risk of age-related macular degeneration. Differences in the severity of an illness or response to treatments may also be manifestations of genetic variations caused by SNPs. For example, two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
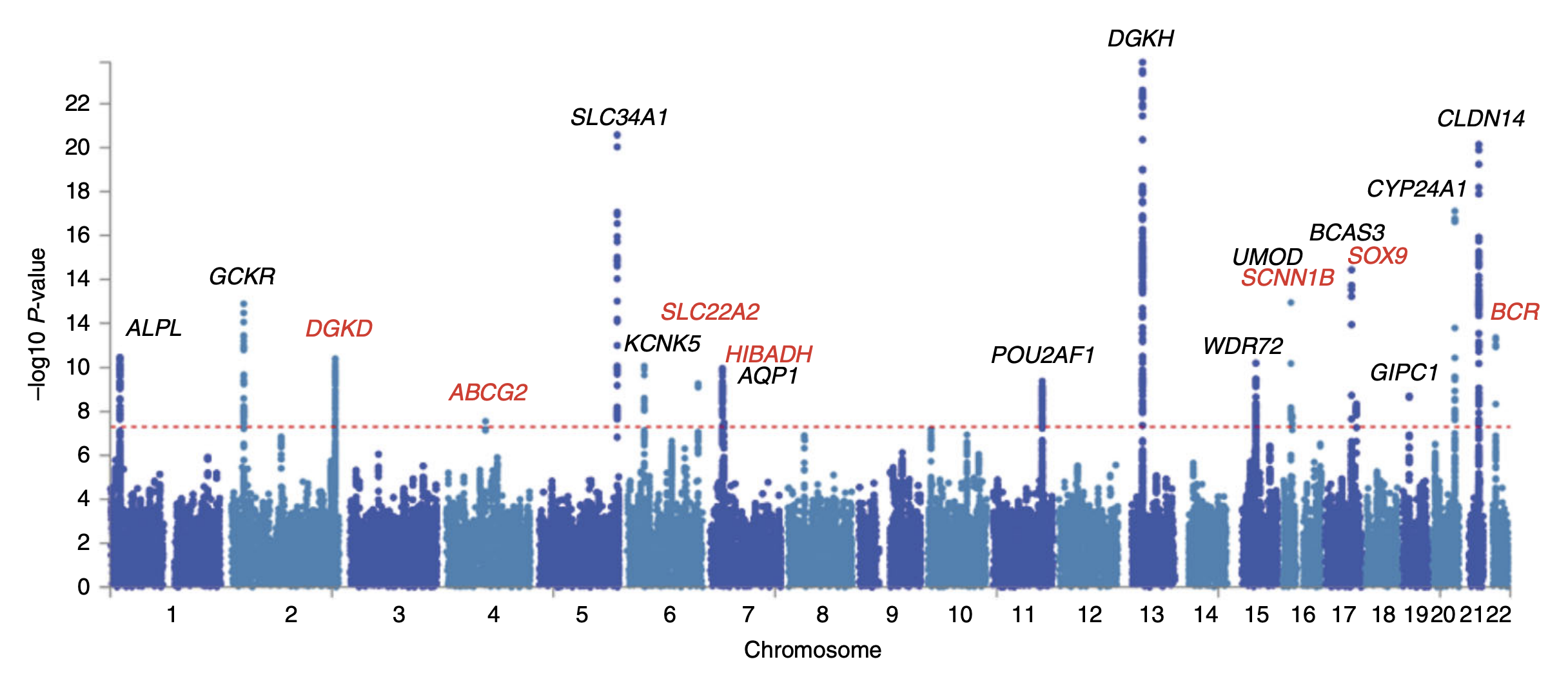
Genome-wide Association Study
In genomics, a genome-wide association study (GWA study, or GWAS), is an observational study of a genome-wide set of Single-nucleotide polymorphism, genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms. When applied to human data, GWA studies compare the DNA of participants having varying phenotypes for a particular trait or disease. These participants may be people with a disease (cases) and similar people without the disease (controls), or they may be people with different phenotypes for a particular trait, for example blood pressure. This approach is known as phenotype-first, in which the participants are classified first by their clinical manifestation(s), as opposed to Genotype-first approach, genotype-first. Each person gives a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MECOM
MDS1 and EVI1 complex locus protein EVI1 (MECOM) also known as ecotropic virus integration site 1 protein homolog (EVI-1) or positive regulatory domain zinc finger protein 3 (PRDM3) is a protein that in humans is encoded by the ''MECOM'' gene. EVI1 was first identified as a common retroviral integration site in AKXD murine myeloid tumors. It has since been identified in a plethora of other organisms, and seems to play a relatively conserved developmental role in embryogenesis. EVI1 is a nuclear transcription factor involved in many signaling pathways for both coexpression and coactivation of cell cycle genes. Gene structure The EVI1 gene is located in the human genome on chromosome 3 (3q26.2). The gene spans 60 kilobases and encodes 16 exons, 10 of which are protein-coding. The first in-frame ATG start codon is in exon 3. mRNA A large number of transcript variations exist, encoding different isoforms or chimeric proteins. Some of the most common ones are: * EVI_1a, EVI_1b, EVI ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
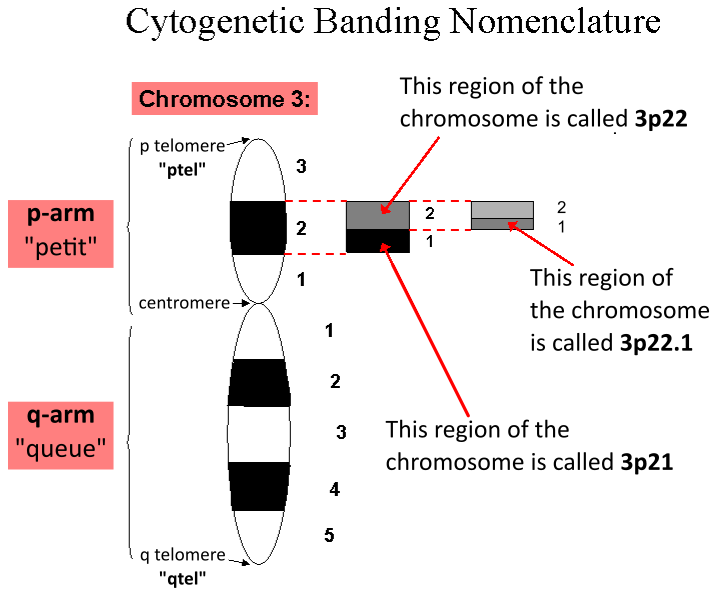
Locus (genetics)
In genetics, a locus (: loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of Human genome#Coding sequences (protein-coding genes), protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000. Genes may possess multiple variants known as alleles, and an allele may also be said to reside at a particular locus. Diploid and polyploid cells whose chromosomes have the same allele at a given locus are called homozygote, homozygous with respect to that locus, while those that have different alleles at a given locus are called heterozygote, heterozygous. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the specific locus or loci responsible for producing a particular phenotype or biological trait. Association ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Whorl
A whorl ( or ) is an individual circle, oval, volution or equivalent in a whorled pattern, which consists of a spiral or multiple concentric objects (including circles, ovals and arcs). In nature File:Photograph and axial plane floral diagram of Friesodielsia desmoides.jpg, Botanical whorls: sepals, petals, leaves, or branches radiating from a single point (photo of flower of Friesodielsia desmoides, family Annonaceae, juxtaposed with diagram of axial cross-section) File:Anisus septegyrus1pl.jpg, Mollusc whorls: Each complete 360° turn in the spiral growth of the shell of the mollusc Anisus septemgyratus, family Planorbidae. File:Baby hairy head DSCN2483.jpg, A hair whorl is a patch of hair growing in a circular direction around a visible center point. File:Fingerprint Whorl.jpg, In a fingerprint, a whorl is each ridge arranged circularly around a central point on the finger. File:Bovine Bone Sample and 430 times Magnification.jpg, In histopathologic architecture, a wh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quantitative Trait Locus
A quantitative trait locus (QTL) is a locus (section of DNA) that correlates with variation of a quantitative trait in the phenotype of a population of organisms. QTLs are mapped by identifying which molecular markers (such as SNPs or AFLPs) correlate with an observed trait. This is often an early step in identifying the actual genes that cause the trait variation. Definition A quantitative trait locus (QTL) is a region of DNA which is associated with a particular phenotypic trait, which varies in degree and which can be attributed to polygenic effects, i.e., the product of two or more genes, and their environment. . These QTLs are often found on different chromosomes. The number of QTLs which explain variation in the phenotypic trait indicates the genetic architecture of a trait. It may indicate that plant height is controlled by many genes of small effect, or by a few genes of large effect. Typically, QTLs underlie continuous traits (those traits which vary continuou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polygene
A polygene is a member of a group of non- epistatic genes that interact additively to influence a phenotypic trait, thus contributing to multiple-gene inheritance (polygenic inheritance, multigenic inheritance, quantitative inheritance), a type of non-Mendelian inheritance, as opposed to single-gene inheritance, which is the core notion of Mendelian inheritance. The term "monozygous" is usually used to refer to a hypothetical gene as it is often difficult to distinguish the effect of an individual gene from the effects of other genes and the environment on a particular phenotype. Advances in statistical methodology and high throughput sequencing are, however, allowing researchers to locate candidate genes for the trait. In the case that such a gene is identified, it is referred to as a quantitative trait locus (QTL). These genes are generally pleiotropic as well. The genes that contribute to type 2 diabetes are thought to be mostly polygenes. In July 2016, scientists reported iden ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |